This project was initiated in the context of the FAIR Data Hackathon organized by NCBI as part of the Bio-IT World conference in Boston in April 2019. If you're not familiar with FAIR, it's a set of principles and protocols for making research resources more Findable, Accessible, Interoperable and Reusable.
The immediate goal is to build on existing open-source tools to make it easier to generate synthetic sequence data for testing pipelines, training, sharing methods and increasing the reproducibility of published studies that involve access-restricted datasets. On the longer term, we are looking to collaborate on developing this further into a proper resource that researchers can use off the shelf.
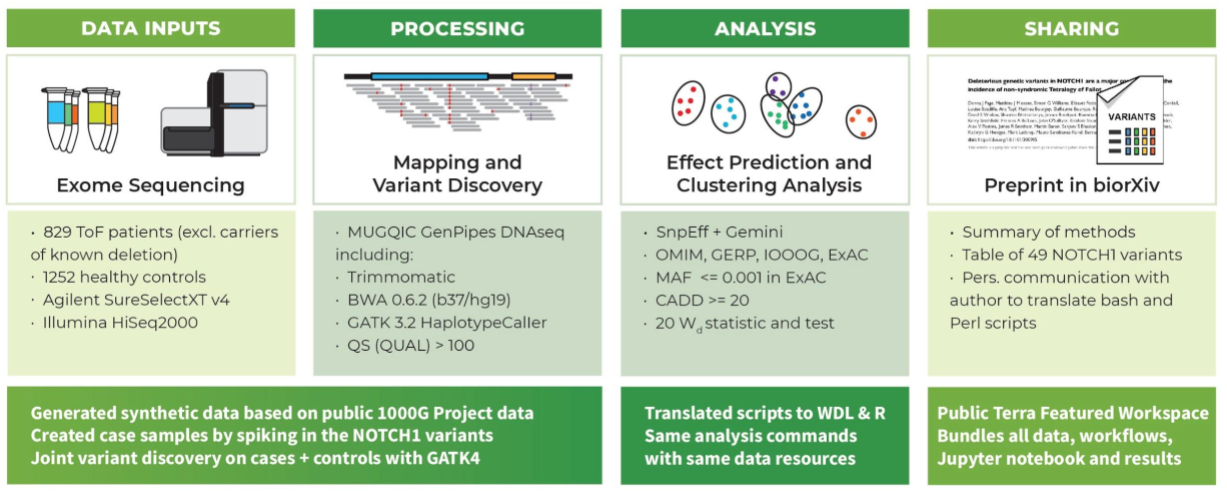
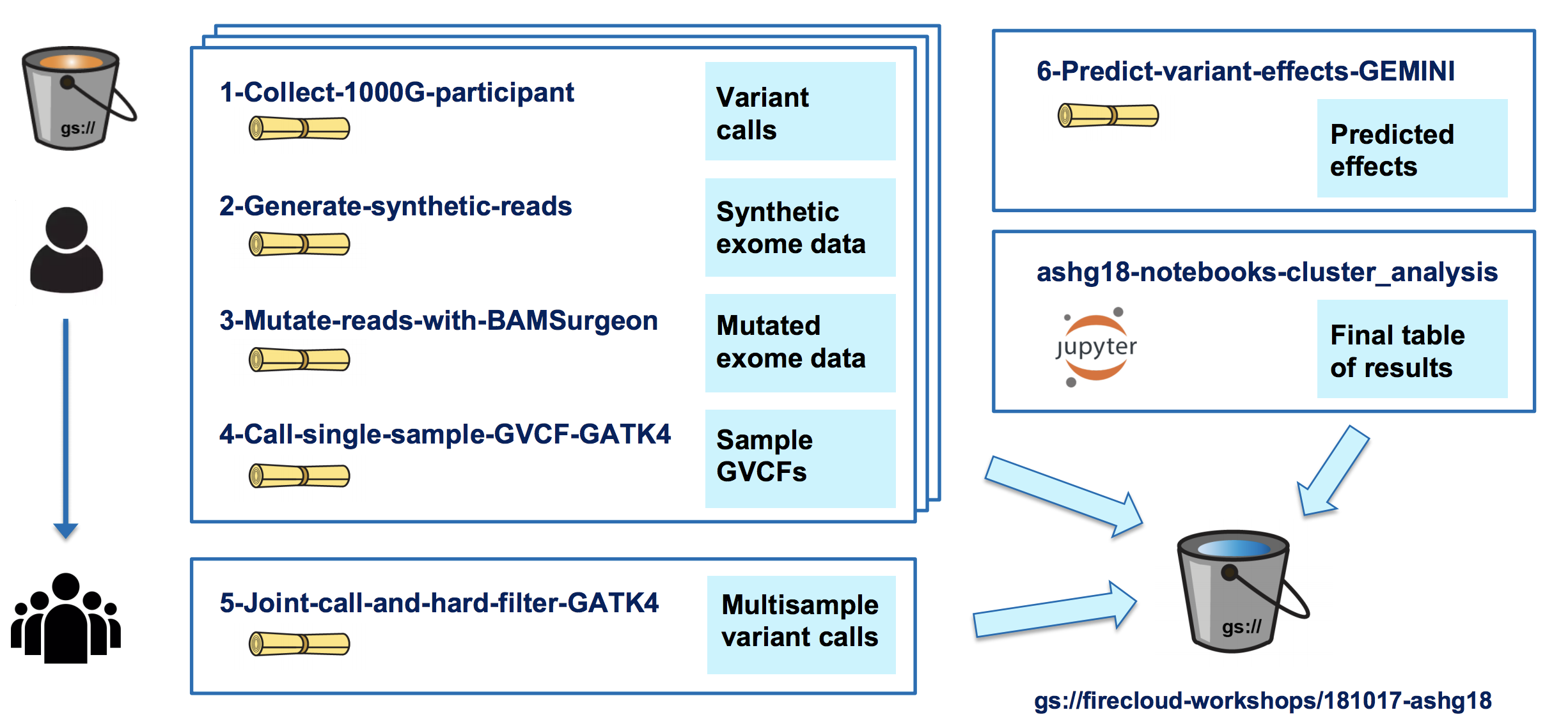
Our team built the first prototype for the ASHG (American Society of Human Genetics) 2018 annual meeting as a demonstration of how to make a reproducible research study. In the original study, the authors identified risk factors for congenital heart disease using exome data from a cohort of more than 800 patients. To make this study reproducible, we needed to generate a synthetic data set, since the original data were private (we decided that it would be simpler than making the original data set sufficiently anonymous). We started from publicly available VCFs from the 1000 Genomes Project, generated synthetic exomes and spiked in mutations of interest. As part of that project, we wrote some pipelines to leverage existing tools (including NEAT and BamSurgeon) for generating synthetic data. You can find a poster that summarizes the project here, and the Terra workspace is here.
Here's an infographic that breaks down the main components of the original paper and the work that was necessary to reproduce it:
And here's an overview of the workflows and notebook we implemented:
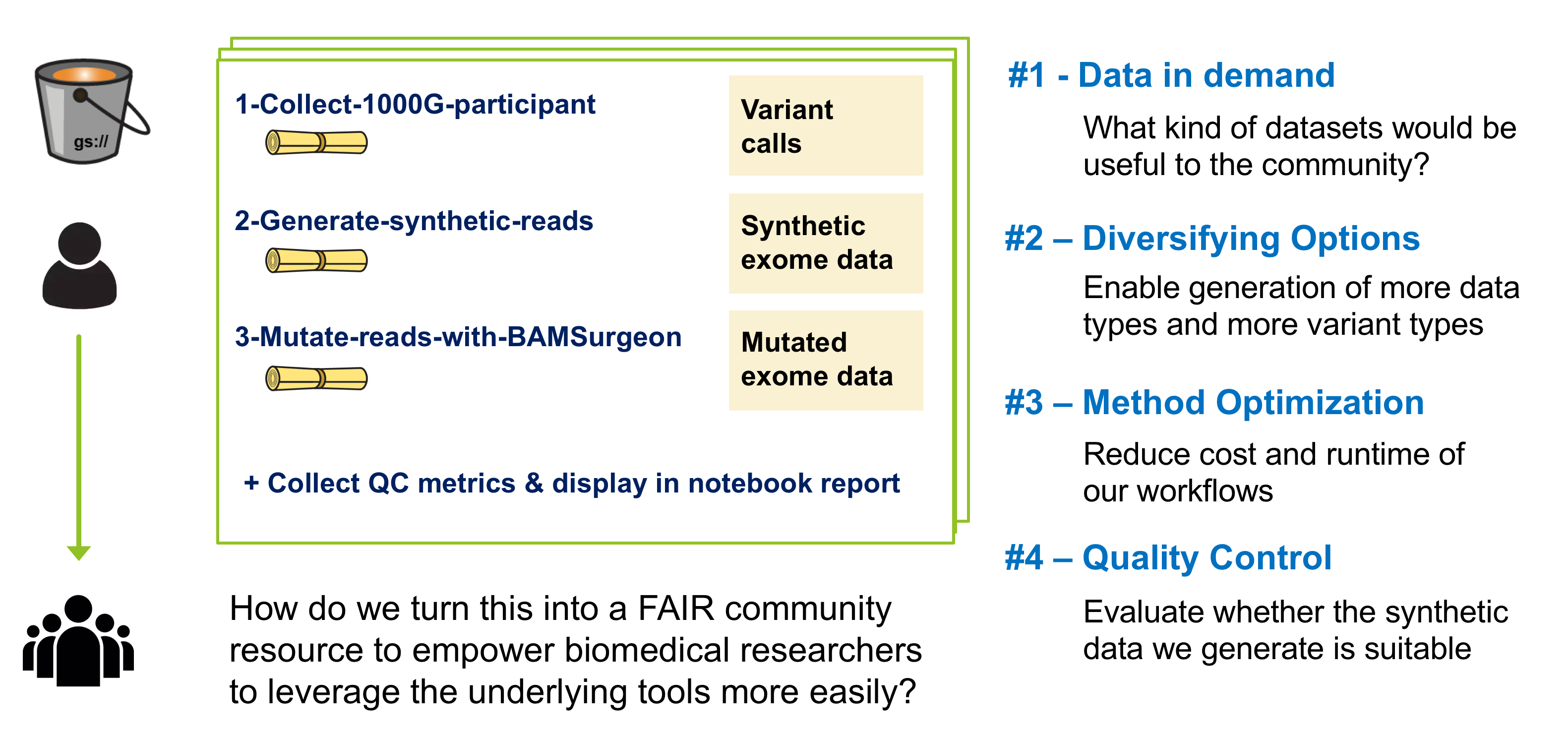
Creating an accurate synthetic dataset of that size for the original project was a fairly painful process, and we realized there would be great value in turning our prototype into a community resource. We decided to pursue this work through various hackathons as well as use it as a case study.
Terra workspace
Github repository
Here's an overview of the four main buckets of work we pursued:
We summarized our results here on Day 2, and later presented a more polished recap and summary in the FAIR data track of the BioIT conference. Those slides are here in PPT and PDF, and on slideshare here.
TODO: Summarize work and results
The following people have all contributed to this project in some form even though some may not show up in the Github records.
TODO: Add Helsinki workshop participants who wish to be listed
Ernesto Andrianantoandro
Dan Rozelle
Jay Moore
Rory Davidson
Roma Kurilov
Vrinda Pareek
Eric Banks
Dan Billings
Bhanu Gandham
Allie Hajian
Anton Kovalsky
Ruchi Munshi
Tiffany Miller
Adelaide Rhodes
Andrey Smirnov
Geraldine Van der Auwera