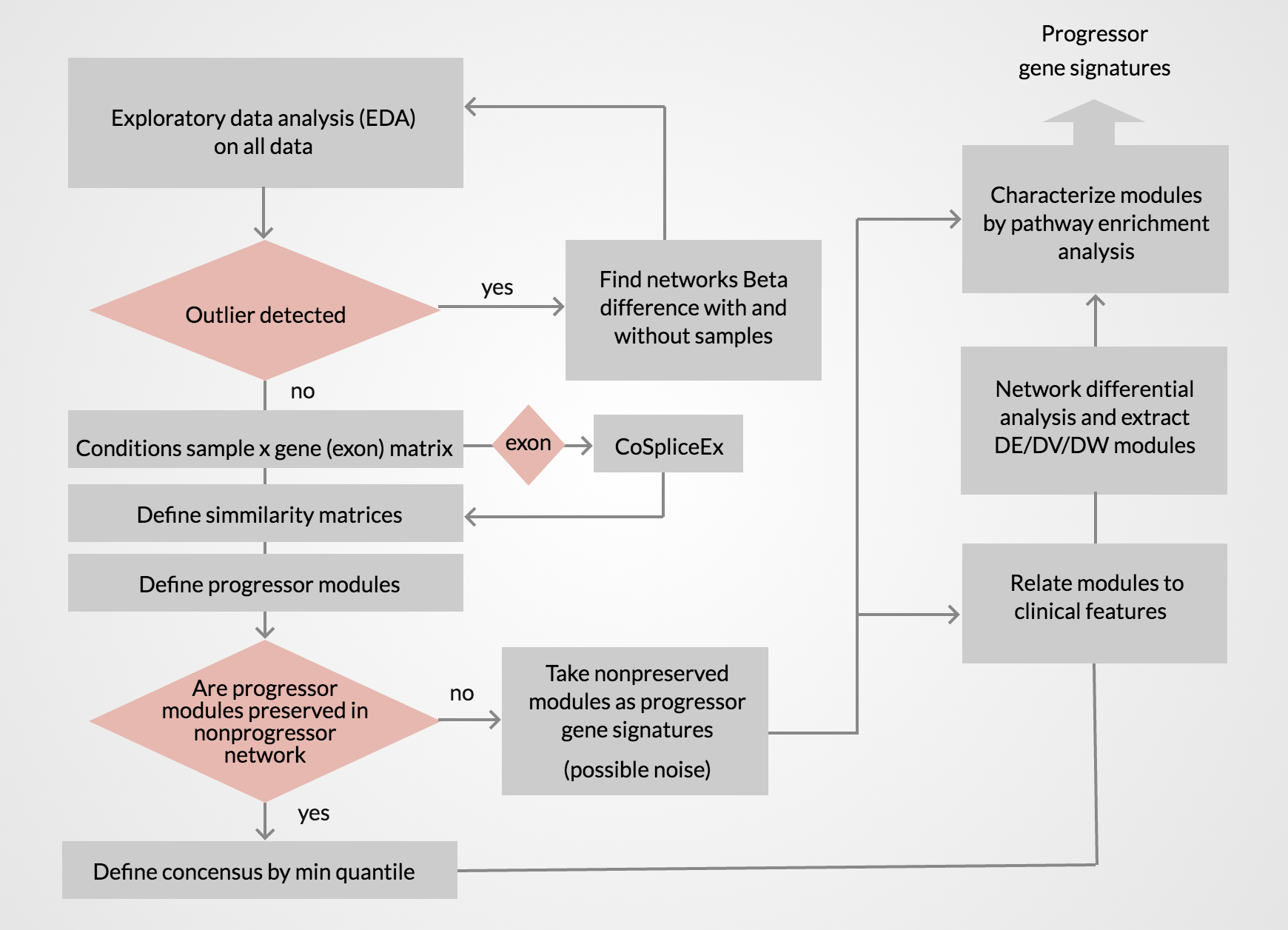
Coexpression and CoSpliceEx weighted network analysis of Head and Neck Squamous Cell Carcinoma (HNSCC).
We analyzed 229 patients’ samples from The Cancer Genome Atlas (TCGA) previously annotated by Bornstein et al (2016) on their progression status.
https://www.frontiersin.org/articles/10.3389/fgene.2018.00183
https://digitalcommons.ohsu.edu/etd/3824/
R version 3.3.1 or 3.3.2
- Ovidiu Dan Iancu open access research and code
- WGCNA
- Bornstein et al. study (2016)
- TCGA
- Hadley Wickham Data Science, Data import, and R software engineering libraries
This code was written over a short period of time to complete educational milestones and should not be taken as a computationally automated process/pipeline. However, each code segment (between code comments) may be repurposed. The publication requires this code to be public.