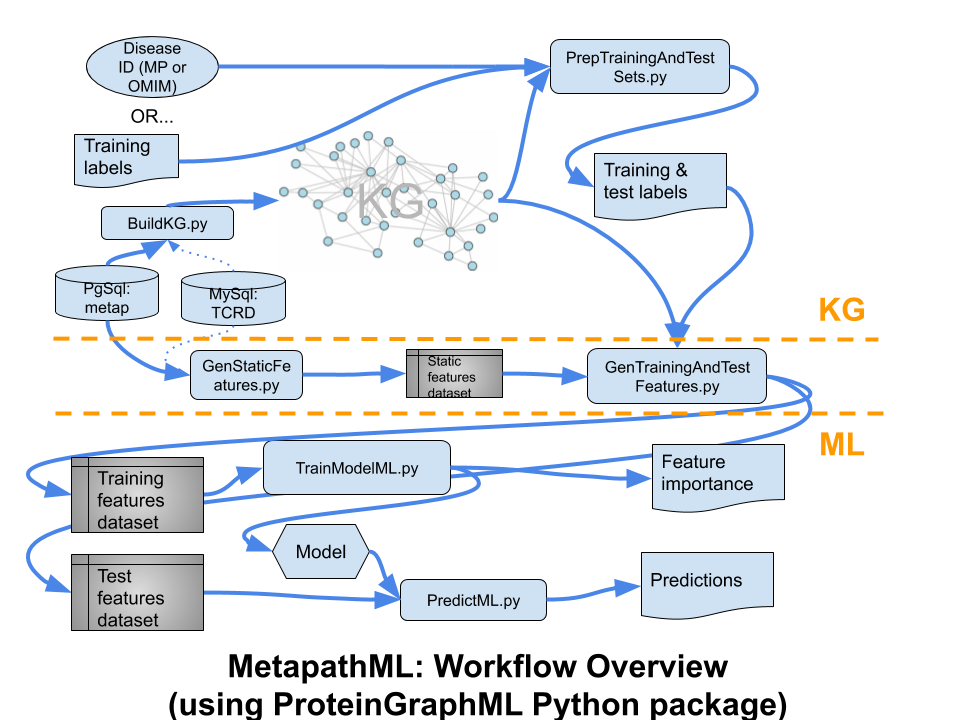
This software is designed to to predict disease-to-protein (protein-coding gene) associations, from a biomedical knowledge graph, via machine learning (ML). This codebase abstracts the ML from the domain knowledge and data sources, to allow reuse for other applications. The input PostgreSQL relational database is converted to a knowledge graph, then converted to feature vectors by metapath matching, based on an input disease, defining a training set of proteins. Then XGBoost is used to generate and optimize a predictive model.
- Dependencies
- How to Run Workflow
- Also see
- ComparingResults (Python vs R)
- Visualization (In development.)
- R 3.5+
- R packages:
data.table,Matrix,RPostgreSQL - Python 3.4+
- Python packages:
xgboost,scikit-learn,networkx,pandas,pony,matplotlib,xlrd,XlsxWriter - PostgreSQL database
metap.- Edit
DBcreds.yamlwith valid db credentials. Needed throughout workflow.
- Edit
- Hardware: Some of the programs need 20+ GB RAM and Xgboost takes several minutes to train and test the data if the number of CPU cores is less than 8. So, a machine with 20+ GB RAM and 8+ CPU core is preferable for running these codes.
The command-line programs of ProteinGraphML must be executed in the following order. However, the BuildKG and StaticFeatures are one-time steps, re-useable for multiple ML models. Re-run only required if database updated.
BuildKG.py, from the relational db, generates a knowledge graph,
a ProteinDiseaseAssociationGraph, saved as a pickled networkX graph.
Via the adapter and pony
object-relational model (ORM), nodes and edges are queried from the db to comprise the
graph.
Command line parameters:
--o: Pickled KG file.--db: database (olegdb or tcrd) to use to build KG (default: tcrd).--logfile: KG log file (optional).--cyjsfile: Save KG as CYJS file. (optional).--graphmlfile: Save KG as graphML. (optional).--tsvfile: Save KG as TSV file. (optional).
Example commands:
BuildKG.py -h
BuildKG.py --db tcrd --o ProteinDisease_GRAPH.pkl
BuildKG.py --db tcrd --o ProteinDisease_GRAPH.pkl --logfile ProteinDisease_GRAPH.log --cyjsfile ProteinDisease_GRAPH.cyjs --tsvfile ProteinDisease_GRAPH.tsv
GenStaticFeatures.py generates files for static features: lincs, hpa, gtex, and ccle for use by GenTrainingAndTestFeatures.py. Static features are not dependent on training set labels, only the database,
so the same TSV files can be reused for all models, and only needs to be re-run if
the database changes. Note: Requires large memory server, approximately 100GB+,
80GB for this process.
Command line parameters:
--db: database (olegdb or tcrd) to use to generate static features (default: tcrd)--outputdir: output folder to save TSV files.--sources: static features (default: ["gtex", "lincs", "ccle", "hpa"]).--decimals: decimal place for the values (default:3)
GenStaticFeatures.py --db tcrd --source "gtex,hpa" --outputdir . (only gtex and hpa)
GenStaticFeatures.py --db tcrd --outputdir . (for all 4 static features)
PrepTrainingAndTestSets.py generates two files:
- A
pickleed Python dictionary that contains protein_ids for both class 'True' and 'False'. This training set file is needed for running ML for a disease defined by a custom labeled training set, rather than a Mammalian Phenotype (MP) term ID. The custome labeled training set may reference proteins viaprotein_ids or gene symbols; if gene symbols, this code fetches the correspondingprotein_idfor each symbol from the database. The prepared, picked training set usesprotein_ids. - A
pickleed test set of protein_ids, unlabeled, defining the predictions of interest, for testing the trained model.
Command line parameters:
--i: Input file that contains protein_ids/symbols and labels for a given disease, with extension (csv|txt|xlsx|rds).--symbol_or_pid: "symbol" or "pid" (default: symbol).--use_default_negatives: Use default negatives, ~3500 genes with known associations but not with query disease. If false, input training set must include negatives.--db: database (olegdb or tcrd) (default: tcrd)
If the file is a spreadsheet, the header should have "Protein_id Label" or "Symbol Label" and the sheet name should be "Sheet1". If the file is a text file, the Protein_id/symbol and Label should be comma-separated. There should not be any header in the text file. If a disease is not present in the graph, use the corresponding RDS file in this program to generate sets of training and predict protein ids. E.g. 104300.rds, PS168600.rds
Example commands:
PrepTrainingAndTestSets.py -h
PrepTrainingAndTestSets.py --i data/diabetes_pid.txt --symbol_or_pid 'pid' --db tcrd
PrepTrainingAndTestSets.py --i data/autophagy.xlsx --db tcrd
PrepTrainingAndTestSets.py --i data/diabetes.xlsx --use_default_negatives --db tcrd
PrepTrainingAndTestSets.py --i data/Asthma.rds --db tcrd
To generate metapath features from the KG, use GenTrainingAndTestFeatures.py. From the KG
and hard coded metapath patterns, plus the positively labeled proteins in the
training set, feature vectors are generated for all training cases and optionally
predict cases. Normally, any human proteins not in the labeled training set
will be in the predict set. Metapath-based features
must be generated for each model (unlike static features), since how metapath
semantic patterns match the KG depends on the query disease. Mammalian Phenotype ID can also be used
with this program to generate training and predict data sets for ML models.
Command line parameters:
--disease: Mammalian Phenotype ID, e.g. MP_0000180 (Diseases without MP_TERM_ID may give error. So, use their RDS files to create sets of training and predict protein ids usingPrepTrainingAndTestSets.py)--trainingfile: pickled training set, e.g. "diabetes.pkl"--predictfile: pickled predict set, e.g. "diabetes_test.pkl"--outputdir: directory where train and test data with features will be saved, e.g. "diabetes_no_lincs"--kgfile: input pickled KG (default: "ProteinDisease_GRAPH.pkl")--static_data: (default: "gtex,lincs,ccle,hpa")--static_dir: directory of static features files: lincs.tsv, hpa.tsv, gtex.tsv, and ccle.tsv--db: database (olegdb or tcrd) (default: tcrd)
Example commands:
GenTrainingAndTestFeatures.py -h
GenTrainingAndTestFeatures.py --trainingfile data/ATG.pkl --predictfile data/ATG_predict.pkl --outputdir results/ATG --kgfile ProteinDisease_GRAPH.pkl --static_data "gtex" --static_dir . --db tcrd
GenTrainingAndTestFeatures.py --disease MP_0000180 --outputdir results/MP_0000180 --kgfile ProteinDisease_GRAPH.pkl --static_data "gtex,lincs,ccle,hpa" --static_dir . --db tcrd
GenTrainingAndTestFeatures.py --trainingfile data/PS118220.pkl --predictfile data/PS118220_predict.pkl --outputdir results/PS118220 --kgfile ProteinDisease_GRAPH.pkl --static_data "gtex,lincs,ccle,hpa" --static_dir . --db tcrd
TrainModelML.py, from the training set feature vectors, or a training set
implicated by specified disease (Mammalian Phenotype ID),
executes the specified ML procedure, training a predictive model, then saved to a
reusable file (.model). The procedure XGBGridSearch uses
XGBoost, trains a model with cross-validation and grid-search parameter optimization,
generates a list of important features used by the classification model.
Command line parameters:
PROCEDURE(positional parameter):XGBGridSearch: Grid search for optimal XGBoost parameters.XGBCrossValPred: 5-fold cross-validation, one iteration.XGBKfoldsRunPred: 5-fold cross-validation, multiple iterations. In each iteration, data is randomly divided into train and test set (80:20). Model trained on train set is tested on test set. The average, min and max AUC are computed using the classification results of test data.
--trainingfile: Training set file, produced byGenTrainingAndTestFeatures.py.--resultdir: directory for output results--nrounds_for_avg: number of iterations to compute average AUC, accuracy, and MCC. This is used for procedureXGBKfoldsRunPred.--rseed: random seed that XGBoost should use for procedureXGBGridSearch(default:1234)--nthreds: number of CPU threads for procedureXGBGridSearch(default:1).--xgboost_param_file: XGBoost configuration parameter file (e.g. XGBparams.txt). This is used forXGBCrossValPredandXGBKfoldsRunPred. XGBparams.txt created by GridSearch can be used for this parameter. Modify XGBparams.txt if any parameter needs to be changed.--db: database (olegdb or tcrd) (default: tcrd)--static_data: (default: "gtex,lincs,ccle,hpa")--static_dir: directory of static features files: lincs.tsv, hpa.tsv, gtex.tsv, and ccle.tsv
Example commands:
TrainModelML.py -h
TrainModelML.py XGBGridSearch --trainingfile results/ATG/ATG_TrainingData.pkl --rseed 1234 --nthreads 32 --resultdir results/ATG --db tcrd --static_data "gtex,lincs,ccle,hpa" --static_dir .
TrainModelML.py XGBCrossValPred --trainingfile results/ATG/ATG_TrainingData.pkl --resultdir results/ATG --xgboost_param_file XGBparams.txt --db tcrd --static_data "gtex,lincs,ccle,hpa" --static_dir .
TrainModelML.py XGBKfoldsRunPred --trainingfile results/ATG/ATG_TrainingData.pkl --resultdir results/ATG --xgboost_param_file XGBparams.txt --nrounds_for_avg 5 --db tcrd --static_data "gtex,lincs,ccle,hpa" --static_dir .
Results will be saved in the specified --resultsdir. See logs for specific subdirectories and output files, including:
- Saved XGBoost model (.model).
- Feature importance lists (.tsv, .xlsx).
PredictML.py, Using the model trained on the training set and KG, predicts the probability of True class for
proteins. The procedure XGBPredict uses the saved XGBoost model, and generates results for predictions on all proteins in the test set.
Command line parameters:
PROCEDURE(positional parameter):XGBPredict: load the saved model
--modelfile: trained model (e.g. results/autophagy_test20191003/XGBCrossVal.model).--predictfile: predict data file, produced byPrepTrainingAndTestSets.py(e.g. "diabetesPredictData.pkl")--resultdir: directory for output results--infofile: protein information file with full path. The file should contain tdl, fam, uniprot data.--db: database (olegdb or tcrd) (default: tcrd)
Example commands:
PredictML.py -h
PredictML.py XGBPredict --predictfile results/ATG/ATG_predict_PredictData.pkl --model results/ATG/XGBCrossValPred.model --resultdir results/ATG --db tcrd --infofile data/plotDT.xlsx
Results will be saved in the specified --resultsdir. See logs for specific subdirectories and output files, including:
- Predictions with probabilities for all proteins (.tsv, .xlsx).
- The code currently assumes that all nodes are unique, that proteins are integer IDs, and the only ints in the graph.
- New data sources can be supported by adding new Adapter class in
ProteinGraphML/DataAdapter/. - New ML procedures may be added to
ProteinGraphML/MLTools/Procedures/.
Workflow overview diagram: