The blood-brain barrier (BBB) protects and regulations the microvasculature in the central nervous system (CNS) by inhibiting the transportation or passage of toxins and pathogens from the blood. Because of its resistance to exogenous compounds, the BBB also poses a challenge for the delivery of neuroactive molecules (i.e. drugs) into the CNS. Understanding small molecules' BBB permeability is therefore vital for CNS drug discovery, and should be considered at an early stage in the drug-development pipeline to avoid costly late-stage failures.
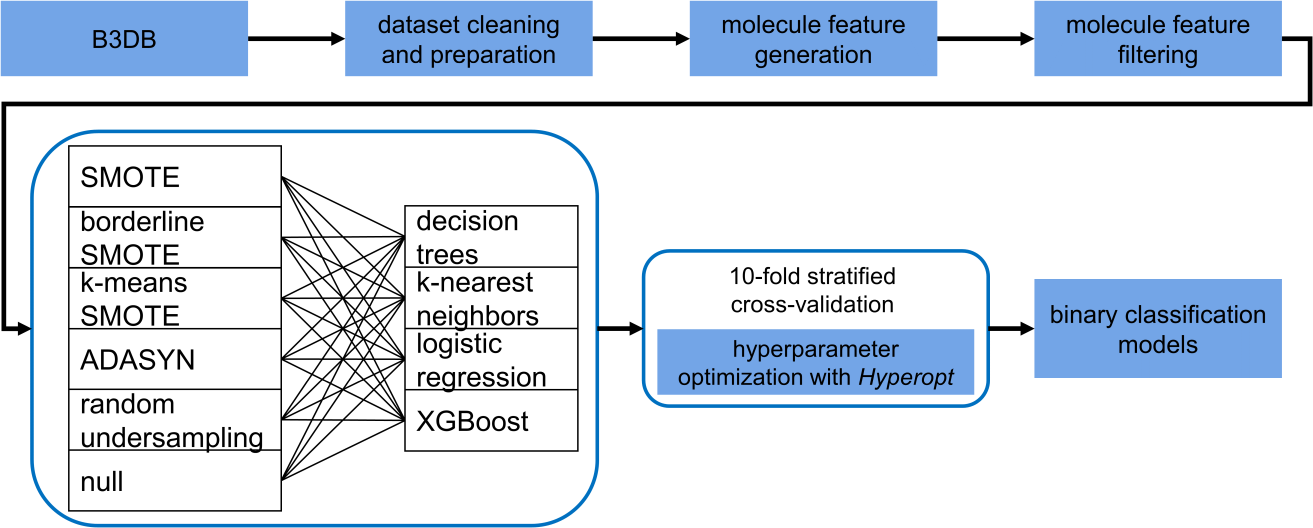
B3clf uses data from 7407 molecules, taken from our curated dataset,
B3DB. It supports
24 different models, with four different classification algorithms (dtree for decision
trees, logreg for logistical regression, knn for KNN, xgb for XGBoost) and six resampling
strategies (classic_RandUndersampling, classic_SMOTE, borderline_SMOTE, k-means_SMOTE, classic_ADASYN,
and common for no resampling), as shown below.
The workflow of B3clf is summarized in the following diagram:
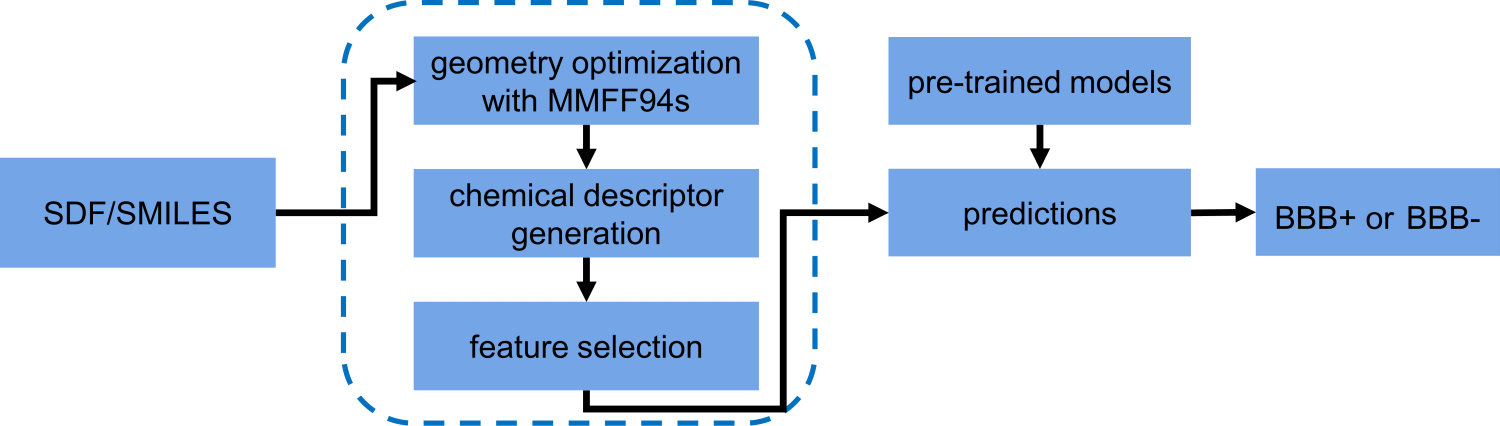 The output is the molecule name/ID, the predicted probability, and the BBB
permeability label. The predicted probability makes it easy to benchmark our models, enabling
calculations of ROC, precision-recall curves, etc..
The output is the molecule name/ID, the predicted probability, and the BBB
permeability label. The predicted probability makes it easy to benchmark our models, enabling
calculations of ROC, precision-recall curves, etc.. b3clf has been tested on Windows 10,
Linux and MacOS.
It is recommended to work with a virtual environment with Python >=3.7. Here is the code
snippet that can be used to install our package.
# create a virtual environment with conda
conda create -y -n b3clf_py37 python=3.7
# or
# conda env create --file environment.yml
conda activate b3clf_py37
# download B3clf
git clone git@github.com:theochem/B3clf.git
cd B3clf
# install rdkit
conda install -c rdkit rdkit>=2020.09.1.0
# install other dependencies
# pip install -r requirements.txt
# or with
# conda install --file requirements_conda.txt
# install B3clf package
pip install .Last but not least, Java 6+ is required in order to compute chemical descriptors with
padelpy which is a python wrapper of
Padel.
Once can easily get the help document from bash with
b3clf --helpwhich prints out,
usage: b3clf [-h] [-mol MOL] [-sep SEP] [-clf CLF] [-sampling SAMPLING]
[-output OUTPUT] [-verbose VERBOSE]
[-keep_features KEEP_FEATURES] [-keep_sdf KEEP_SDF]
b3clf predicts if molecules can pass blood-brain barrier with resampling
strategies.
optional arguments:
-h, --help show this help message and exit
-mol MOL Input file with descriptors.
-sep SEP Separator for input file. Default="\s+|\t+".
-clf CLF Classification algorithm type. Default=xgb.
-sampling SAMPLING Resampling method type. Default=classic_ADASYN.
-output OUTPUT Name of output file, CSV or XLSX format.
Default=B3clf_output.xlsx.
-verbose VERBOSE If verbose is not zero, B3clf will print out the
predictions. Default=1.
-keep_features KEEP_FEATURES
To keep computed feature file ("yes") or not ("no").
Default=no.
-keep_sdf KEEP_SDF To keep computed molecular geometries ("yes") or not
("no"). Default=no.
In Python, it is also doable with
from b3clf import b3clf
b3clf?There are two ways to use B3clf for BBB predictions, as a command-line (CLI) tool and as a Python
package.
Now B3clf supports SMILES and SDF text files. Three example files are provided in the
test sub-folder. A simple usage example is:
b3clf -mol test_input_sdf.sdf -clf xgb -sampling classic_ADASYN -output test_SMILES_pred.xlsx -verbose 1which outputs
| ID | B3clf_predicted_probability | B3clf_predicted_label | |
|---|---|---|---|
| 0 | H1_Bepotastine | 0.142235 | 0 |
| 1 | H1_Quifenadine | 0.981108 | 1 |
| 2 | H1_Rupatadine | 0.967724 | 1 |
Please use the following citation in any publication using B3clf:
Fanwang Meng, et al, Blood-Brain Barrier Permeability Predictions of
Organic Molecules with XGBoost and Resampling Strategies, Journal, page, volume, year, doi.The B3DB database is an essential ingredient in B3clf:
@article{Meng_A_curated_diverse_2021,
author = {Meng, Fanwang and Xi, Yang and Huang, Jinfeng and Ayers, Paul W.},
doi = {10.1038/s41597-021-01069-5},
journal = {Scientific Data},
number = {289},
title = {A curated diverse molecular database of blood-brain barrier permeability with chemical descriptors},
volume = {8},
year = {2021},
url = {https://www.nature.com/articles/s41597-021-01069-5},
publisher = {Springer Nature}
}For any suggestions or questions, post questions in GitHub Discussion Board.
- Add link to manuscript
- Add CITATION.cff once manuscript is published
- Add environment.yml for conda to install everything in one step