- Theophil Trippe, Technische Universität Berlin, Institut für Mathematik, Berlin, Germany
- Martin Genzel, Utrecht University, Mathematical Institute, Utrecht, Netherlands
- Jan Macdonald, Technische Universität Berlin, Institut für Mathematik, Berlin, Germany
- Maximilian März, Technische Universität Berlin, Institut für Mathematik, Berlin, Germany
Our approach to the Helsinki Deblur Challenge 2021 (see: https://www.fips.fi/HDC2021.php#anchor1) is to train end-to-end deblurring neural networks, i.e., in the evaluation phase the deblurred images are directly estimated by the networks without the need of solving an optimization problem.
The network architecture is a slight modification (more down- and up-sampling steps, GroupNormalization [1], more aggressive sub-sampling for an increased field-of-view) of the standard U-Net [2]. This model was originally proposed for image segmentation and now also forms a prominent backbone for data-driven reconstruction schemes for imaging inverse problems [3,4,5].
Such powerful network architectures may adapt very well to specific data, but possibly "overfit" to the provided text images of the challenge (i.e. the network would always output text regardless of the input image). Such a behavior would violate the goal of the challenge, which is to identify general purpose methods capable of deblurring also other types of images.
A canonical way to address this aspect is an integration of model-based knowledge into the reconstruction pipeline, e.g., in the form of unrolled networks. We did not pursue this strategy for two reasons: (a) In our experience, unrolled networks typically only show a clear advantage for inverse problems that involve a domain change (e.g. images -> sinograms in CT). (b) In our experience, unrolled networks typically only outperform if a rather precise forward model is known.
Still following the philosophy of incorporating model-based knowledge, we instead pursue the following strategy: (a) We estimate the forward model from the given training data. A key component is to model lens distortion effects in addition to blurring. (b) We use this model to simulate additional training data (text & other images).
This approach has three advantages: (a) Including other images in the simulated training data allows us to pass the challenge "sanity check". (b) Increasing the text training data beyond the provided 100 examples per font per blur level improves the final performance of the networks. (c) Accounting for the radial distortion renders the problem approximately translation invariant, which is beneficial for a subsequent processing by the U-Nets.
In more detail, our training proceeds in three phases:
(I) We use a division-model for the radial lens distortion after the convolution with the blurring kernel. The three parameters of the distortion and the 701 x 701 unknown parameters of the blurring kernel are jointly estimated from the training data via empirical risk minimization and PyTorch's automatic differentiation. It turns out that there is a "mustache" type distortion and, as expected, a disk-like convolution kernel.
(II) The first step of the reconstruction pipeline is an inversion of the radial distortion using the estimated parameters from (I). The subsequent U-Nets are pre-trained on a random stream of simulated data of random text symbols and other "sanity check" images.
(III) In a final phase, we fine-tune on the provided challenge data. In order not to "lose" the generalization to "sanity check" images from (II) we keep a small fraction of simulated data in the training.
For all blur levels we use the same parametric model of the forward operator and network architecture, but train each level separately.
- We have included two YAML files
environment_cuda.ymlandenvironment_nocuda.ymlfrom which our Conda environment can be reconstructed using the commands
conda env create -f environment_cuda.ymlor
conda env create -f environment_nocuda.ymlrespectively. The former is intended for a use with Cuda (GPU support), the latter for a use without Cuda (no GPU support).
- Download the network weights from https://1drv.ms/u/s!ApMVkVzMgNK3gocTlXCBFUCJoIoyLQ?e=HHOart and place them in the
HDC_weightsdirectory (alternatively you can specify theWEIGHTS_PATHvariable inconfig.pyto point to another directory (relative or absolute path))
- Activate the Conda environment via
conda activate hdc_tub- To test if the installation and setup was successful run
python demo.py <output-path>This should generate results comparable to the example text images shown below.
- For the main evaluation run
python main.py <input-path> <output-path> <blur-step>This should run on a GPU if there is one available and on the CPU otherwise.
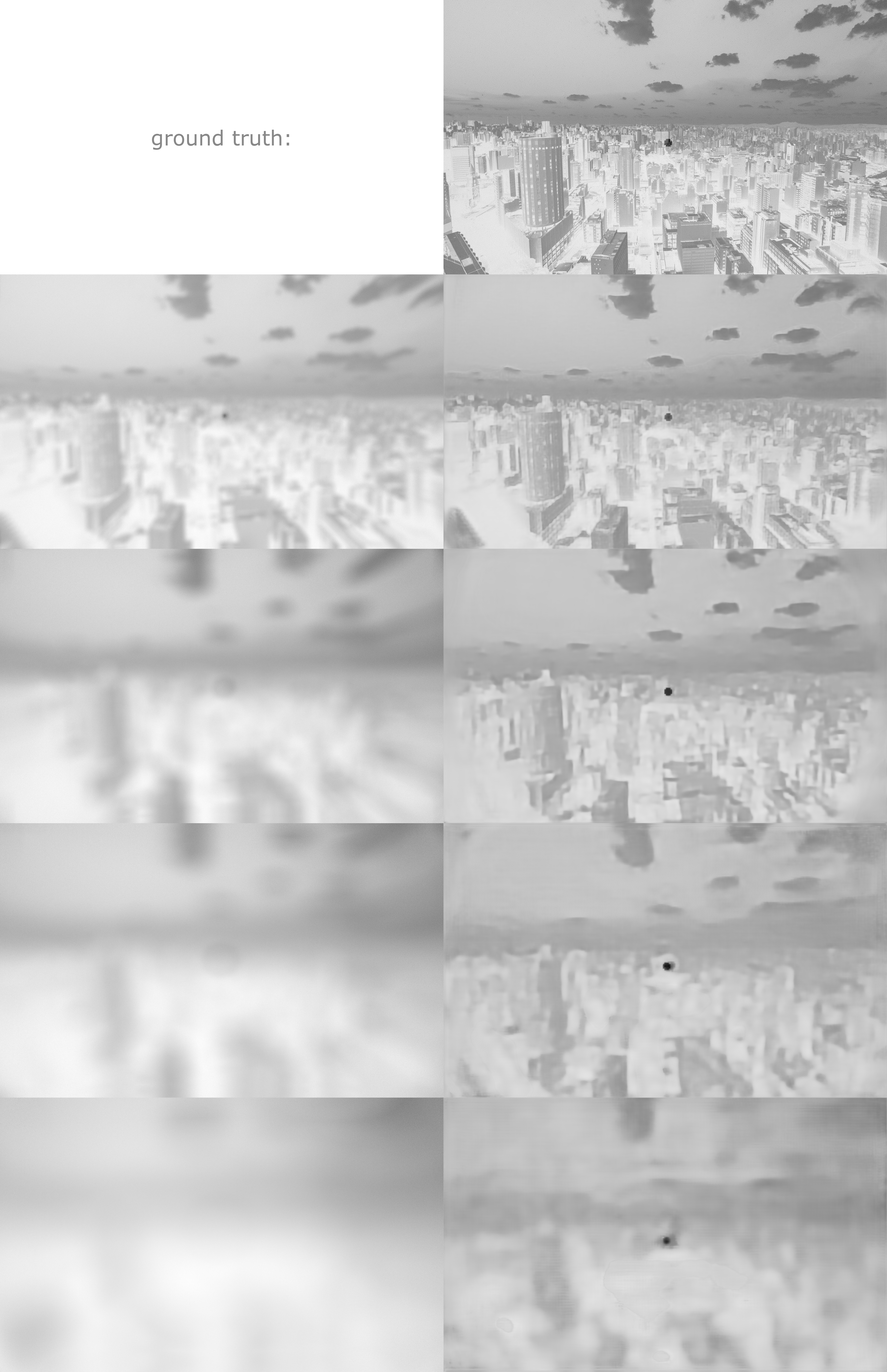
To verify that everything is working correctly we provide example reconstructions of a text image and a "sanity check" image. Both were held back and not seen by the network during training. The "sanity check" has an artifact of our forward model simulation in the center. This will not happen for real data.
Our implementation of the U-Net is based on and adapted from https://github.com/mateuszbuda/brain-segmentation-pytorch/.
The "sanity check" images are taken from https://synthia-dataset.net/.
[1] Wu, Yuxin, and Kaiming He. "Group normalization." Proceedings of the European conference on computer vision (ECCV). 2018.
[2] Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
[3] Jin, Kyong Hwan, et al. "Deep convolutional neural network for inverse problems in imaging." IEEE Transactions on Image Processing 26.9 (2017): 4509-4522.
[4] Kang, Eunhee, Junhong Min, and Jong Chul Ye. "A deep convolutional neural network using directional wavelets for low‐dose X‐ray CT reconstruction." Medical physics 44.10 (2017): e360-e375.
[5] Genzel, Martin, Jan Macdonald, and Maximilian März. "AAPM DL-Sparse-View CT Challenge Submission Report: Designing an Iterative Network for Fanbeam-CT with Unknown Geometry." arXiv preprint arXiv:2106.00280 (2021).