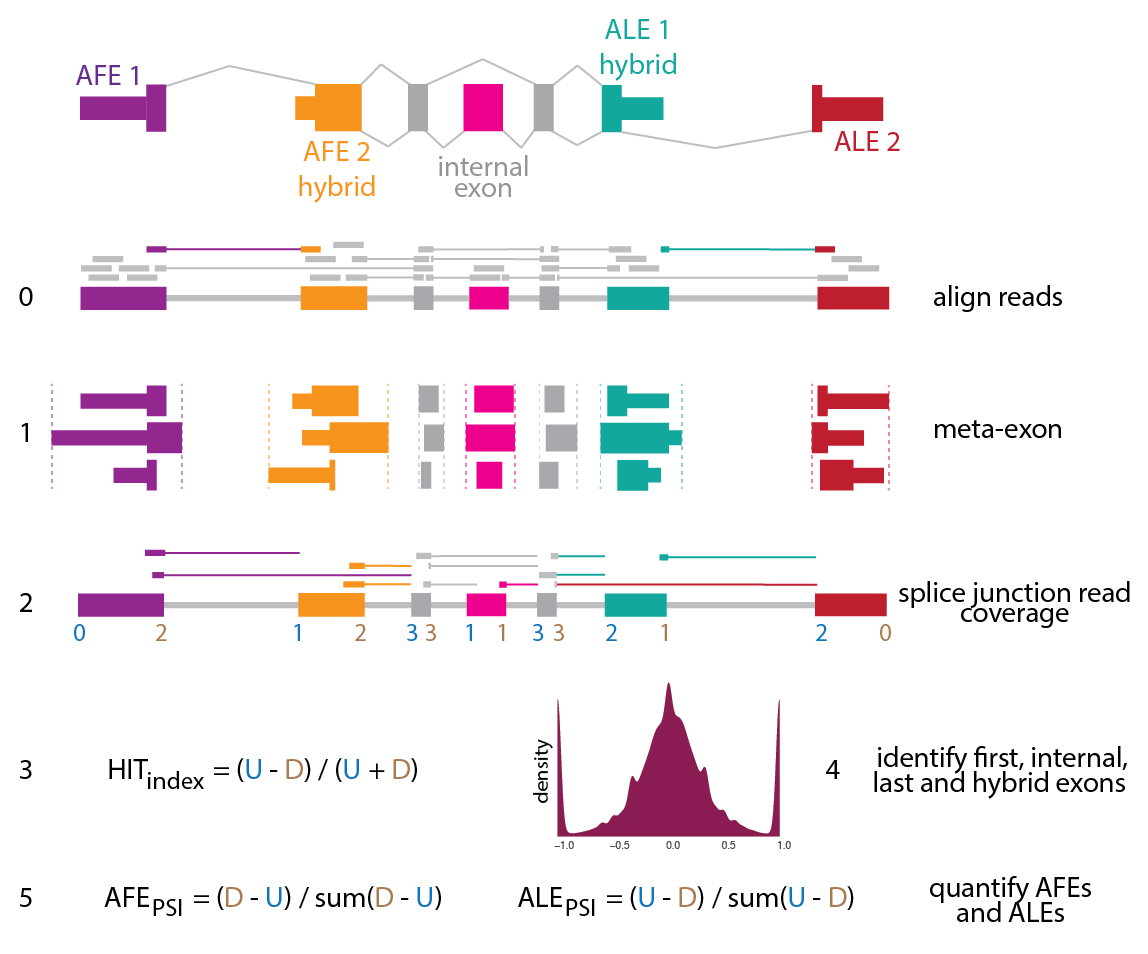
Citation: Fiszbein A, McGurk M, Calvo Roitberg E, Kim GY, Burge CB, and Pai AA. (2021). Widespread occurrence of hybrid internal-terminal exons in human transcriptomes. (bioRxiv) doi: https://doi.org/10.1101/2021.05.27.446076
The HITindex is a pipeline to classify hybrid, internal, or terminal exons from RNA-seq data by modeling ratios of splice junction coverage. The pipeline involves two major scripts, which can be run independently:
- HITindex_annotate: Annotate metaexons from a gtf file by collapsing overlapping consituent exons.
- HITindex_classify: Calculate HIT index metrics and classify metaexons into one of 5 exon-types: first, first-internal, internal, internal-last, and last exons.
- samtools (v1.3)
- bedtools (v2.26.0)
- python (v3.6.3)
- scipy (v1.5.2)
- numpy (v1.19.2)
- pysam (v0.16)
- pybedtools (v0.8.1)
- pandas (v0.25.3)
- pymc3 (v3.9.3)
The HITindex was designed to be run on bam files containing mapped reads. Here, we describe the usage of the two main steps of the HITindex pipeline. Below, we provide sections that discuss alternative parameter usage for classification and quantification, as well as a tutorial walking through all the steps necessary to run the HITindex.
Annotate metaexons from a gtf file by collapsing overlapping consituent exons. This step includes (a) annotating how often a constituent exon is used as a first, internal, or last exon in annotated isoforms, (b) saving the coordinates of each constituent exons, and (c) adding buffer regions in which to associate junction reads with an exon.
usage: HITindex_annotate.py [-h] --gtf gtf [--reverse] [--ss3buffer] [--ss5buffer]
--outfile output
optional arguments:
-h, --help show this help message and exit
Input:
--gtf gtf gtf to be indexed (default: None)
Parameters:
--reverse use if exons are sorted by transcriptional direction rather than by reference
coordinate (default: False)
--ss3buffer intronic buffer region included upstream of 3ss of exon for counting
reads. suggested = 50nt. (default: 0)
--ss5buffer intronic buffer region included downstream of 5ss of exon for counting
reads. suggested = 20nt. (default: 0)
Output:
--outfile output name for output bed with merged/annotated exons (default: None)
Calculate HIT index metrics and classify metaexons into one of 5 exon-types: first, first-internal, internal, internal-last, and last exons. This step includes (a) calculating the HITindex and generative model metrics, (b) flagging exons likely affected by edge effects, (c) classifying exons, and (d) calculating PSI values for alternative first and last exon usage.
usage: HITindex_classify.py [-h] [--junctionReads] [--HITindex] [--identifyTerminal] [--calculatePSI]
--outname output [--bam] [--juncbam] [--readtype {single,paired}]
[--readstrand {fr-unstrand,fr-firststrand,fr-secondstrand}] [--bed] [--overlap]
[--readnum] [--metrics] [--parameters] [--bootstrap] [--metricsID] [--edge]
optional arguments:
-h, --help show this help message and exit
--junctionReads Extract junction reads (default: False)
--HITindex Calculate HITindex (default: False)
--classify Classify terminal, hybrid, and internal exons (default: False)
--calculatePSI Calculate PSI values (default: False)
--outname name of file(s) for final metric. required for everything except --junctionReads. (default: None)
read information:
--bam original bam from which to extract junction reads. required if
--junctionReads (default: None)
--juncbam junction read bam. required if --junctionReads or --HITindex (default:
None)
--readtype {single,paired}
type of read (default: paired)
--readstrand {fr-unstrand,fr-firststrand,fr-secondstrand}
directionality of RNA-seq data (default: fr-firststrand)
exon information:
--bed bed file with merged/annotated exons. Output from HITindex_annotate.py.
required if --HITindex (default: None)
HITindex:
parameters for running HIT index
--overlap overlap of split read with exon region (nt) (default: 10)
--readnum minimum number of reads for confidence in HITindex (sum of R + L) (default:
2)
classify:
information for identifying exon types
--metrics HITindex output file, required if --HITindex is not specified. (default:
None)
--parameters file specifying HITindex and generative model thresholds for classifying
exons. (default: HIT_identity_parameters.txt)
--bootstrap bootstrapping iterations to get confidence intervals and p-values
(default: 1000)
psi:
parameters for calling PSI values
--metricsID HITindex identification output file, required if --classify is not
specified. (default: None)
--edge exclude exons flagged as being affected by the edge effect from PSI
calculations (default: False)
In this tutorial, we walk through all the steps to run the HITindex pipeline, in the minimum number of command lines and each step individually. For each step, we discuss the possible parameters that can be changed, how to do so, and the considerations involved in each of the parameters. Finally, we show example inputs and outputs of each step (with column explanations) so the user knows what to expect and can make custom files as needed.
Step 1: Identify and Annotate Metaexons
Step 2: Extracting Junction Reads
Step 3: Calculating HITindex metrics
Align raw reads in fastq format to the genome with your favorite splicing-aware mapper (ie. STAR | hisat2) to obtain a sorted, indexed bam file. When building a STAR index or running hisat2, we recommend using the same gtf annotation that you will use for downstream steps.
For instance, to map with STAR (using ENCODE parameters) and index the bam:
STAR --outFilterType BySJout --outFilterMultimapNmax 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --outFilterMismatchNmax 999 --outFilterMismatchNoverLmax 0.04 --alignIntronMin 20 --alignIntronMax 1000000 --alignMatesGapMax 1000000 --outSAMtype BAM SortedByCoordinate
samtools index [bamfile].bam
This step takes in an annotation file (gtf file) and outputs a bed file of metaexons after collapsing and annotating overlapping exons.
Example usage:
python HITindex_annotate.py --gtf annotations.gtf --ss3buffer 50 --ss5buffer 20 --outfile metaexons.bed
Types of GTF files
(1) Exons in gtf are sorted by genome coordinates (default):
1 havana gene 11869 14409 . + . gene_id "ENSG00000223972"; gene_name "DDX11L1";
1 havana transcript 11869 14409 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; gene_name "DDX11L1";
1 havana exon 11869 12227 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "1"; gene_name "DDX11L1";
1 havana exon 12613 12721 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "2"; gene_name "DDX11L1";
1 havana exon 13221 14409 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "3"; gene_name "DDX11L1";
1 havana gene 34554 36081 . - . gene_id "ENSG00000237613"; gene_name "FAM138A";
1 havana transcript 34554 36081 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; gene_name "FAM138A";
1 havana exon 34554 35174 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; exon_number "3"; gene_name "FAM138A";
1 havana exon 35277 35481 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; exon_number "2"; gene_name "FAM138A";
1 havana exon 35721 36081 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; exon_number "1"; gene_name "FAM138A";
(2) Exons in gtf are sorted by transcriptional direction (use --reverse):
1 havana gene 11869 14409 . + . gene_id "ENSG00000223972"; gene_name "DDX11L1";
1 havana transcript 11869 14409 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; gene_name "DDX11L1";
1 havana exon 11869 12227 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "1"; gene_name "DDX11L1";
1 havana exon 12613 12721 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "2"; gene_name "DDX11L1";
1 havana exon 13221 14409 . + . gene_id "ENSG00000223972"; transcript_id "ENST00000456328"; exon_number "3"; gene_name "DDX11L1";
1 havana gene 34554 36081 . - . gene_id "ENSG00000237613"; gene_name "FAM138A";
1 havana transcript 34554 36081 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; gene_name "FAM138A";
1 havana exon 35721 36081 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; exon_number "1"; gene_name "FAM138A";
1 havana exon 35277 35481 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; exon_number "2"; gene_name "FAM138A";
1 havana exon 34554 35174 . - . gene_id "ENSG00000237613"; transcript_id "ENST00000417324"; exon_number "3"; gene_name "FAM138A";
Buffer regions around metaexons
Users can chose to add a buffer region around metaexon boundaries within which to associate junction reads to a particular metaexon. This is meant to account for some flexibility in TSS and TES definitions, which are often hard to precisely define and thus less likely to be precise at the single nucleotide level in annotation sets. While the default is set to 0nt for both the 5' and 3' buffer regions, we suggest using --ss3buffer 50 for a 50nt buffer upstream of the 3' splice site (5' end of exon) and --ss5buffer 20 for a 20nt buffer downstream of the 5' splice site (3' end of exon):
Metaexon annotations
Three bed files are output:
(1) Precise metaexon boundaries
(2) Boundaries defined by the user-defined buffer regions
(3) Associating metaexon names with the coordinates of the constitutent overlapping exons that were combined to create the metaexon
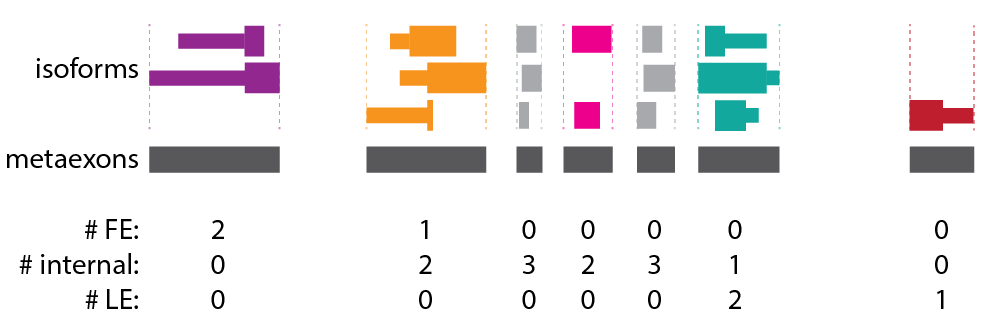
The 4th "name" column of all three bed files will include the original metaexon coordinates, the gene name (from the gtf), and a 'TXPT' annotation, which is the total number of isoforms for that gene. The first two bed files include additional information in the 4th column about how many times the constituent exons are first (FE), internal, or last exons (LE) within annotated isoforms from the gtf file (or appear as single exon isoforms), as shown below. For this example, the 4th column will include TXPT:3, because there are 3 isoforms in the toy gene.
Example metaexon bed output:
chr16 15703135 15704123 chr16:15703135-15704123;ENSG00000133392;TXPT:12;FE:0;internal:0;LE:6;singleexon:0 0 -
chr16 15708803 15708841 chr16:15708803-15708841;ENSG00000133392;TXPT:12;FE:0;internal:4;LE:0;singleexon:0 0 -
chr16 15711113 15711270 chr16:15711113-15711270;ENSG00000133392;TXPT:12;FE:1;internal:0;LE:0;singleexon:0 0 -
chr16 15712890 15715081 chr16:15712890-15715081;ENSG00000133392;TXPT:12;FE:0;internal:5;LE:2;singleexon:0 0 -
chr16 15715164 15715272 chr16:15715164-15715272;ENSG00000133392;TXPT:12;FE:0;internal:7;LE:0;singleexon:0 0 -
chr16 15717140 15719308 chr16:15717140-15719308;ENSG00000133392;TXPT:12;FE:0;internal:19;LE:0;singleexon:0 0 -
Example buffer bed output (--ss3buffer 20 & --ss5buffer 50:
chr16 15703115 15704173 chr16:15703135-15704123;ENSG00000133392;TXPT:12;FE:0;internal:0;LE:6;singleexon:0 0 -
chr16 15708783 15708891 chr16:15708803-15708841;ENSG00000133392;TXPT:12;FE:0;internal:4;LE:0;singleexon:0 0 -
chr16 15711093 15711320 chr16:15711113-15711270;ENSG00000133392;TXPT:12;FE:1;internal:0;LE:0;singleexon:0 0 -
chr16 15712870 15715131 chr16:15712890-15715081;ENSG00000133392;TXPT:12;FE:0;internal:5;LE:2;singleexon:0 0 -
chr16 15715144 15715322 chr16:15715164-15715272;ENSG00000133392;TXPT:12;FE:0;internal:7;LE:0;singleexon:0 0 -
chr16 15717120 15719358 chr16:15717140-15719308;ENSG00000133392;TXPT:12;FE:0;internal:19;LE:0;singleexon:0 0 -
Example constituent bed output:
chr16 15703135 15704123 chr16:15703135-15704123;ENSG00000133392;TXPT:12;15703135-15704123,15703172-15704123,15703172-15704123,15703741-15704123,15703787-15704123,15703991-15704123 -
chr16 15708803 15708841 chr16:15708803-15708841;ENSG00000133392;TXPT:12;15708803-15708836,15708803-15708841,15708803-15708841,15708838-15708841 0 -
chr16 15711113 15711270 chr16:15711113-15711270;ENSG00000133392;TXPT:12;15711113-15711270 0 -
chr16 15712890 15715081 chr16:15712890-15715081;ENSG00000133392;TXPT:12;15712890-15715081,15712896-15715081,15714909-15715081,15714909-15715081,15714909-15715081,15714909-15715081,15714909-15715081 0 -
chr16 15715164 15715272 chr16:15715164-15715272;ENSG00000133392;TXPT:12;15715164-15715272,15715164-15715272,15715164-15715272,15715164-15715272,15715164-15715272,15715164-15715272,15715164-15715272 0 -
chr16 15717140 15719308 chr16:15717140-15719308;ENSG00000133392;TXPT:12;15717140-15719308,15717140-15717348,15717140-15717348,15717140-15717348,15717140-15717348,15717140-15717348,15717140-15717348,15718315-15718438,15718315-15718438,15718315-15718438,15718315-15718438,15718315-15718438,15718315-15718438,15719220-15719308,15719220-15719308,15719220-15719308,15719220-15719308,15719220-15719308,15719220-15719308 0 -
Example usage using default parameters to run all 4 steps in tandem:
python HITindex_classify.py --junctionReads --bam sample.sorted.bam --juncbam sample.sorted.junctions.bam
--HITindex --bed metaexon.bed_ss3-20ss5-50.buffer
--classify --calculatePSI
--outname sampleHITindex
To only extract junction reads:
python HITindex_classify.py --junctionReads --bam sample.sorted.bam --juncbam sample.sorted.junctions.bam --readtype paired --readstrand fr-firststrand
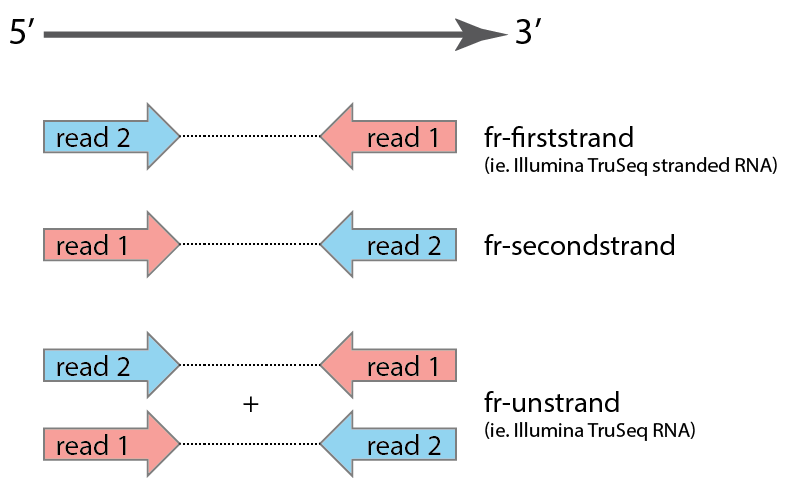
Junction reads are extracted by parsing the CIGAR strings of mapped reads. To correctly assign junction reads the user needs to provide information about read type and strandedness of the reads:
(1) Read type can be changed with --readtype with option {single or paired}, default: paired
(2) Strandedness of the reads can be changed with --readstrand with options {fr-firststrand, fr-secondstrand, fr-unstrand}, default: fr-firststrand
Strandedness is determined by the type of library preparation protocol. We borrow the library strandedness naming convention from Tophat/Bowtie:
Output
This step results in bam files containing only the junction reads, named using --juncbam. These junction bam files can be specified in later steps, without needing to re-run --junctionReads to extract junction reads again.
To only calculate HITindex metrics and run the generative model:
python HITindex_classify.py --HITindex --juncbam sample.sorted.junctions.bam --readtype paired --readstrand fr-firststrand --bed metaexon.bed_ss3-20ss5-50.buffer --overlap 10 --readnum 5 --outname sampleHITindex
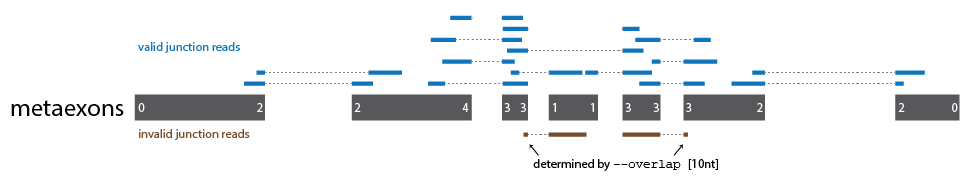
Junction reads are assigned to metaexons based on their overlap with the upstream or downstream boundaries of the metaexon. Note that the junction site does not need to be directly aligned with the exact metaexon boundary coordinates and junction reads are counted regardless of the identity of the connected exon. The minimum overlap length for junction reads is determined by --overlap, with a default of 10nt.
The HITindex and generative model probabilities are calculated for metaexons that have a minimum number of reads as determined by --readnum, with a default of 2 reads. For datasets with sufficient coverage, we recommend using at least 5 reads.
Output
This step results in a .exon file with the following columns:
| Column Name | Description |
|---|---|
| exon | exon name, with coordinates of metaexon |
| gene | gene name (from GTF file) |
| strand | strand |
| nTXPT | total number of annotated isoforms for the gene |
| nFE, nINTERNAL, nLE, nSINGLE | number of times constituent exons of this metaexon are annotated as first, internal, or last exons, or appear as a single exon isoform |
| nUP, nDOWN | number of upstream and downstream splice junction reads |
| HITindex | HITindex |
| dist_to_TSS | metaexon distance to upstream most expressed exon |
| dist_to_TES | metaexon distance to downstream most expressed exon |
| edge | flag indicating whether metaexon is likely to be influenced by edge effects |
| PofF, PofI, PofL | generative model posterior probabilities for first, internal, and last exon classifications |
| PofFI, PofIL | generative model posterior probabilities for hybrid exon classifications |
| downstream_fraction | |
| HIT_postmean |
To only classify exons:
python HITindex_classify.py --classify --metrics sampleHITindex.exon --parameters HIT_identity_parameters.txt --bootstrap 1000
Exons are classified using the .exon file output from the last step and the included HIT_identity_parameters.txt file (reproduced below), which defines the thresholds used across the HITindex metric, statistical confidence metrics, and posterior probabilities from the generative model that are used to classify exons. Users can change these thresholds by changing the values in the HIT_identify_parameters.txt or create custom files with the same format. Custom files can be specified using --parameters.
HIT_identity_parameters file:
# |HITindex| threshold for calling terminal exons [0.0, 1.0]
HITterminal 1.0
# |HITindex| threshold for calling hybrid exons [0.0, 1.0]
HIThybrid 0.3
# bootstrapping p-value threshold for HITindex significance [0.0, 1.0]
HITpval 1
# confidence interval to use for HITindex significance (none, 0.75, 0.95, 0.95)
HIT_CI none
# probability threshold for medium confidence with generative model [0.0, 1.0]
prob_med 0.5
# probability threshold for high confidence with generative model [0.0, 1.0]
prob_high 0.8
Bootstrapping
Bootstrapping is used to calculate a confidence interval and two different p-values related to the HITindex metric. The number of bootstrap iterations used is determined by --bootstrap, with a default of 1000 runs. This is the rate-limiting step for the HITindex pipeline, where reducing the bootstrap n will increase speed, but decrease statistical confidence.
Output
This step adds 7 columns to the existing .exon file:
| Column Name | Description |
|---|---|
| CI_75 | 75% confidence intervals [low,high], using bootstrapped iterations |
| CI_90 | 90% confidence intervals [low,high], using bootstrapped iterations |
| CI_95 | 95% confidence intervals [low,high], using bootstrapped iterations |
| pval_CI | bootstrapping p-value, indicating probability that the confidence interval overlaps the internal exon distribution |
| pval_internal | bootstrapping p-value, indicating probability of observing an internal exon with the same or more extreme HITindex |
| ID | the exon-type classification |
| ID_position | the exon-type classification after accounting for potential edge effect exons |
Users can run this step multiple times with varying thresholds, but since the original file is modified, we suggest duplicating the .exon file for each set of thresholds and then specifying the new .exon files with --metrics.
To only calculate PSI values:
python HITindex_classify.py --calculatePSI --metricsID sampleHITindex.exon --edge --outname sampleHITindex
Percent spliced in (PSI) values are used to quantify relative alternative terminal exon usage. Exons classified as "first", "FirstInternal_medium" or "FirstInternal_high" are used to calculate alternative first exon (AFE) PSI values, while exons classified as “last”, “InternalLast_medium” or “InternalLast_high” are used to calculate PSI values for alternative last exon (ALE) usage. If the user includes the --edge flag, exons flagged as potentially being affected by edge-effects in the ID_position column of the .exon file are not included in the PSI value calculations. Since this step uses the exon classifications in the previous step, it must be run on an .exon file that has the ID and ID_position columns.
Output
This step results in .AFEPSI and .ALEPSI files with the following columns:
| Column Name | Description |
|---|---|
| gene | gene name (from GTF file) |
| exon | exon name, with coordinates of metaexon |
| strand | strand |
| nTXPT | total number of annotated isoforms for the gene |
| nFE or nLE | number of times constituent exons of this metaexon are annotated as first, internal, or last exons, or appear as a single exon isoform |
| nUP, nDOWN | number of upstream and downstream splice junction reads |
| HITindex | HITindex |
| sumR-L or sumL-R | sum of all read biases across all alternative exons |
| AFEPSI or ALEPSI | PSI value for metaexon |
Users can run this step multiple times by using the --metricsID to specify different .exon files or varying the --edge flag, but should make sure to change the --outname for each of these runs.