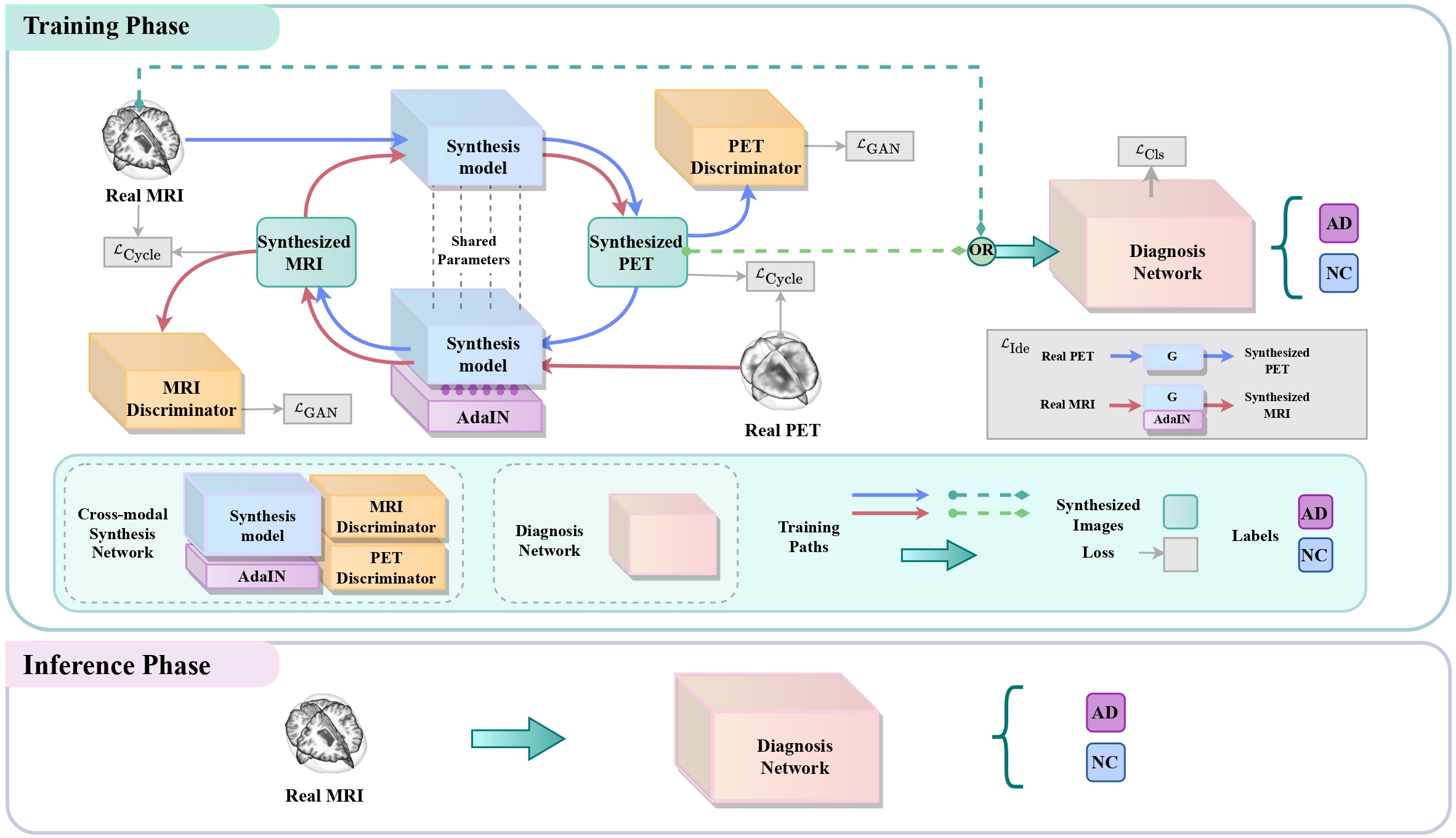
[MedIA 2024] This is a code implementation of the joint learning framework proposed in the manuscript "Joint learning Framework of cross-modal synthesis and diagnosis for Alzheimer's disease by mining underlying shared modality information".[Paper] [Supp.]
🌟🌟🌟 We also plan to open a unified codebase for 3D cross-modality medical synthesis in [code], including updated multi-thread preprocessing steps for MRI and PET, a series of generated methods (CNN-based, GAN-based, and Diffusion-based), and full evaluation pipelines for 3D images.
Among various neuroimaging modalities used to diagnose AD, functional positron emission tomography (PET) has higher sensitivity than structural magnetic resonance imaging (MRI), but it is also costlier and often not available in many hospitals. How to leverage massive unpaired unlabeled PET to improve the diagnosis performance of AD from MRI becomes rather important. To address this challenge, this paper proposes a novel joint learning framework of unsupervised cross-modal synthesis and AD diagnosis by mining underlying shared modality information, improving the AD diagnosis from MRI while synthesizing more discriminative PET images. Additionally, our method is evaluated at the same internal dataset (ADNI) and two external datasets (AIBL and NACC), and the results demonstrated that our framework has good generalization ability.
To diversify the modality information, we propose a novel unsupervised cross-modal synthesis network that implements the inter-conversion between 3D PET and MRI in a single model modulated by the AdaIN module. Here, we present the unsupervised synthesized PET image demo by our ShareGAN after joint training with the downstream diagnosis network.
| Slice direction | Real MRI | Our unsupervised synthesized PET |
Real PET |
|---|---|---|---|
| Axial |  |
 |
 |
| Sagittal | 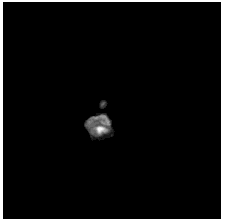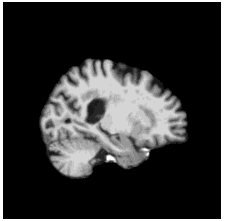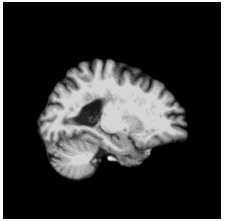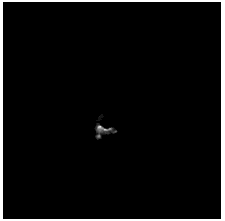 |
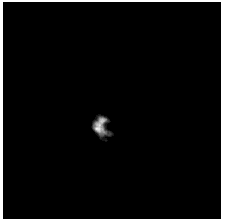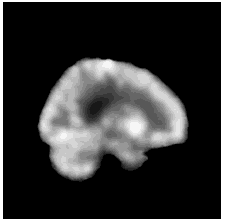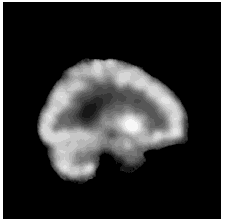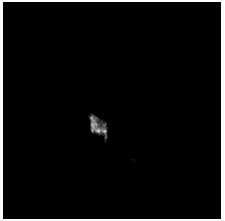 |
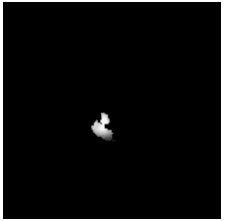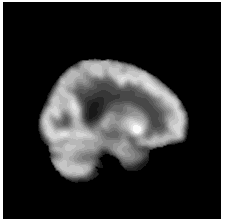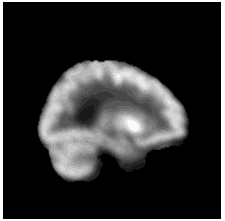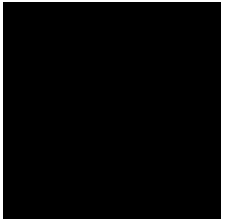 |
| Coronal | 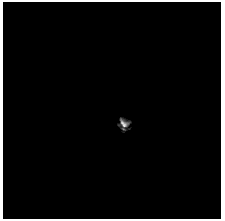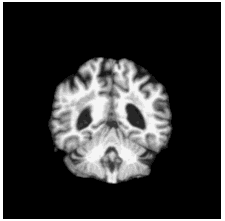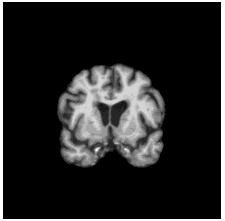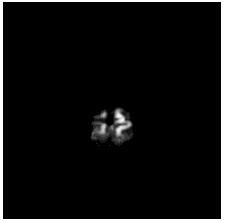 |
 |
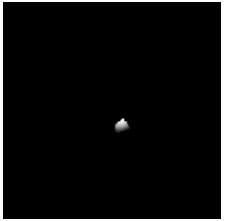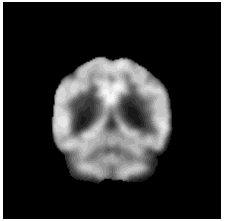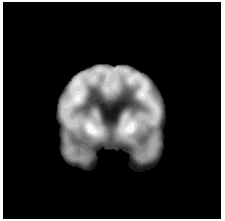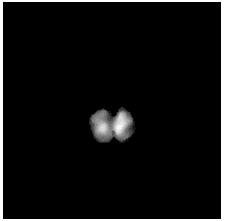 |
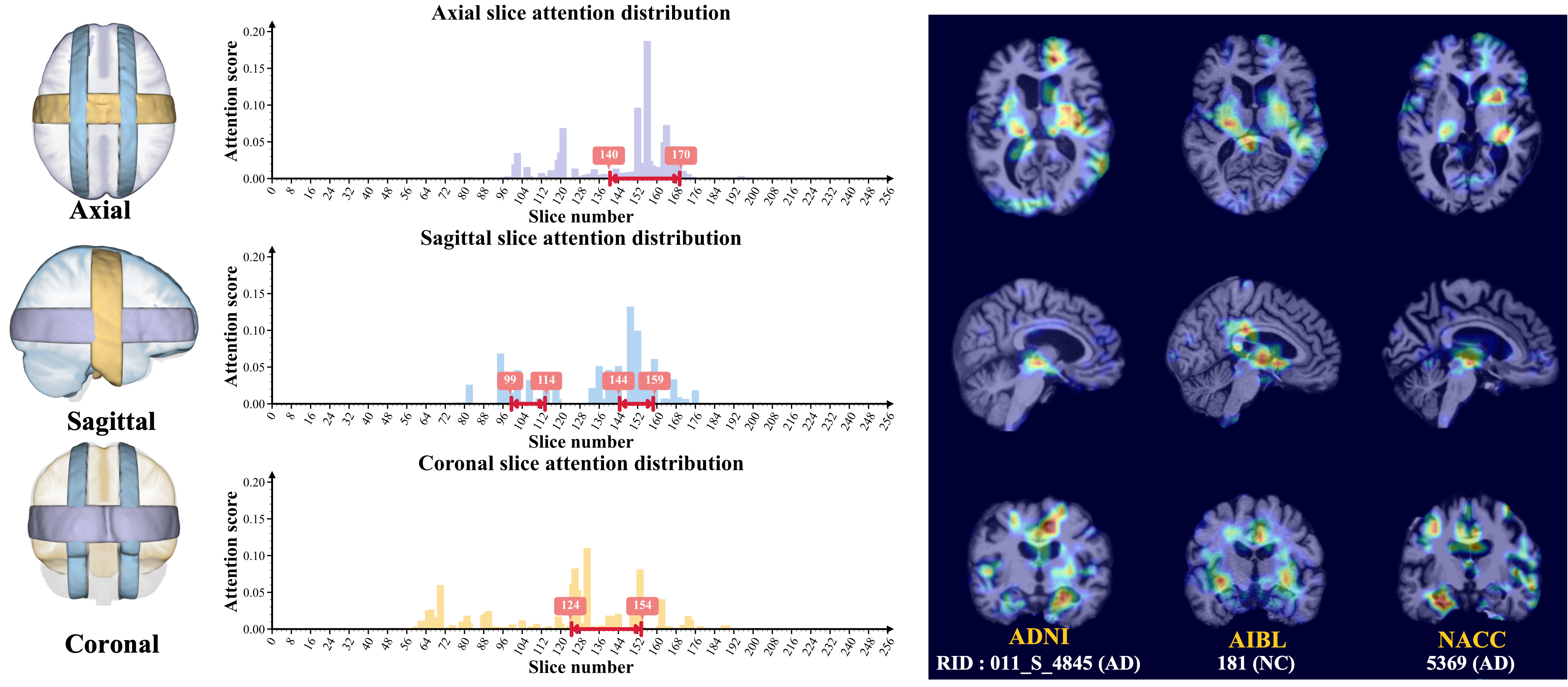
To locate shared critical diagnosis-related patterns, we propose an interpretable diagnosis network based on fully 2D
convolutions, which takes either 3D synthesized PET or original MRI as input. The regions of interest located by our
network are consistent with those associated with AD, such as the ventricles and hippocampus.
First clone the repository:
git clone https://github.com/thibault-wch/Joint-Learning-for-Alzheimer-disease.gitAnd then install other requirements:
pip install -r requirements.txtWe trained, validated and tested the framework using the Alzheimer's Disease Neuroimaging
Initiative (ADNI) dataset. To
investigate the generalizability of the framework, we externally tested the framework on the National Alzheimer's
Coordinating Center (NACC), and the Australian Imaging Biomarkers and Lifestyle Study of
Ageing (AIBL) datasets.
In order to comply with the data requirements of different databases, we have only made the subjectID available
at ./subject_ids.xlsx for visitors to download.
We first use Freesurfer, Flirt and Ants for preprocessing all MRI and PET images (see Supp. B for details). Furthermore,
we transform the preprocessed .nii.gz brain files into .npy format.
In addition, we reorganize and split them into the data pairs for ./utils/UnpairedDataset.py using pickle, the
concrete data pair demo as shown in:
[
( # the MRI file path
'/data/chwang/final_dataset_MRI/lineared/train/0_141_S_0810.npy',
# the PET file path
# (unpaired for ADNI training set, but paired for ADNI validation and test set)
'/data/chwang/final_dataset_PET/lineared/train/0_941_S_4376.npy',
# the diagnosis label of the corresponding MRI subject (NC:0, AD:1)
0),
( '/data/chwang/final_dataset_MRI/lineared/train/1_137_S_0841.npy',
'/data/chwang/final_dataset_PET/lineared/train/0_068_S_4424.npy',
1),
( '/data/chwang/final_dataset_MRI/lineared/train/1_099_S_0372.npy',
'/data/chwang/final_dataset_PET/lineared/train/1_023_S_4501.npy',
1)
...
]Note that the element of external AIBL and NACC testing data pairs only have the MRI file path and the diagnosis label.
We implement the proposed method in the PyTorch library and train them on 2 NVIDIA V100 32G Tensor
Core GPUs.
All the networks are initialized by the kaiming method and trained
using the Adam optimization algorithm with ./scripts.
cd ./scripts/pretraining_stage
sh share_gan_128.shStage 2 : Pre train the synthesis and the diagnosis networks separately with the image size of $256^3$
cd ./scripts/pretraining_stage
# pre-train the cross-modal synthesis network
sh share_gan_256.sh
# pre-train the diagnosis network
sh diag_pretrain.shFirst, move the weights of the pre-trained cross-modal and diagnosis networks to the directory stored for the joint training process. Make sure to rename them with the prefix 'pretrained'. Then, execute the provided shell script.
cd ./scripts/joint_stage
sh joint_learning.shWe also provide the inference codes for the synthesis model and diagnosis network.
cd ./scripts
# for synthesis model
sh test_G.sh
# for diagnosis network
sh test_Cls.shJoint_Learning_for_Alzheimer_disease
├─ components
│ ├─ AwareNet <our diagnosis network>
│ ├─ ShareSynNet <our synthesis model>
│ └─ ...
├─ models
│ ├─ joint_gan_model <our joint learning framework>
│ ├─ share_gan_model <our cross-modal synthesis network>
│ └─ ...
├─ options (different options)
├─ scripts (different stages' scripts)
├─ utils
│ ├─ UnpairedDataset <our defined dataset>
│ ├─ earlystop
│ └─ ...
├─Diag_pretrain.py
├─Diag_test.py
├─Frame_train.py
├─Frame_test.py
├─README.md
├─requirements.txt
├─subject_ids.xlsx
-
We gratefully thank the ADNI, AIBL, and NACC investigators for providing access to the data.
-
Our code is inspired by TSM,Restormer, pytorch-CycleGAN-and-pix2pix,and SwitchableCycleGAN.
If you find this work useful for your research, please 🌟 our project and cite our paper :
@article{wang2024joint,
title = {Joint learning framework of cross-modal synthesis and diagnosis for Alzheimer’s disease by mining underlying shared modality information},
author = {Wang, Chenhui and Piao, Sirong and Huang, Zhizhong and Gao, Qi and Zhang, Junping and Li, Yuxin and Shan, Hongming and others},
journal = {Medical Image Analysis},
volume = {91},
pages = {103032},
year = {2024},
publisher = {Elsevier}
}