Markov is an optimized simulation algorithm written in C for arbitrary Reaction-Diffusion-Systems (RDS).
It simulates the Gillespie algorithm in the thermodynamical limit, and it has been proven [2] that there is no way to do a faster, exact simulation of the trajectory of the RDS.
A RDS is a lattice of reaction cells. The diffusion and reaction have to be chosen, that a diffusion step to a neighbour cell is much more probable than a reaction step in a cell.
The algorithm selects the next step by dividing the cells into reactivity classes based on the dual logarithm of the reaction probability.
This guarantees only a few classes and a relative homogeneous reaction probability inside the class, where a von-Neuman rejection algorithm is used to select a cell with an 75% efficiency.
Inside a cell standard Gillespie can be used.
First selecting the class, then the cell inside the class the algorithm has been proven to be the fastest method possible for this class of problems.
-
The math starts with the Master equation: https://en.wikipedia.org/wiki/Master_equation
-
http://www.worldscientific.com/doi/abs/10.1142/S0129183195000216
You need a full download of Sage http://sagemath.org/ to run this example. You also need some development tools be active, like gcc. I have run this on Ubuntu 12.04.4, and it should run out of the box.
I have split the simulation into the random generator, the standard algorithm, the sage glue code and the domain specific code.
- createtable.c
- gauss55.h
- rand55.h
- random55.h
- rand55.c
- rand55static.c
- logclass.c
- logclass.h
- logclassevent.h
- logclasstest.c
This was the algorithm I developed for my diploma thesis in 1988-1989
- sagemarkov.h
- sagemarkov.c
- markovian.spyx
everything is defined in
- topology.h
- topology.c
The first example is a simple 1-dimensional random walk without reactions
- model.h
- randomwalk.h
call sage from anywhere, the following example also works from the notebook() in a browser
%sage
Sage Version 6.1.1, Release Date: 2014-02-04
sage: os.curdir='/path/to/your/sage-markov'
sage: load_attach_path(os.curdir)
sage: %attach "markovian.spyx"
# no '%' sign in the notebook()
Compiling ./markovian.spyx...
# start a demo
sage: m=Markovian.demo()
100 cells empty initialized
# run a million steps
sage: m.run(1000000)
24.201266334790212
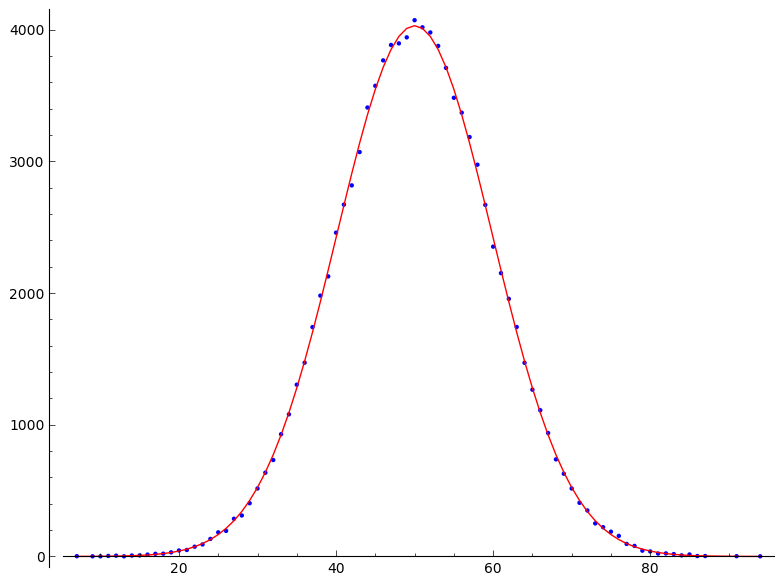
# plot the distribution
sage:m.plot()+m.analytic_plot()another demo, the time series shows a semi log plot of the reactivity, which is in this simple example the same as the number of random walkers
sage: m.destroy()
0
sage: m=Markovian.demo()
100 cells empty initialized
sage: list_plot_semilogy(m.value_by_time(fun=m.reactivity))With
sage: m.reinit()
you can easily reinit your system to the initial state.