This repository implements two different custom KNN algorithms: A simple, yet memory efficient exhaustive search with quadratic runtime but linear memory. Secondly, for multiple queries of the same point cloud, a custom KD-Tree operator in CUDA (GPU), or C++ (CPU). The KD-Tree is always generated using the CPU, but is automatically transferred to the GPU for Tensorflow operations there. The KD-Tree implementation has logarithmic runtime and will quickly
The algorithms' dimensions are currently defined through template parameters and must be known at compile-time therefore. The present version compiles the library for the dimensionalities 1, 2, 3.
int main()
{
std::vector<test_T> point_cloud; //KD-Tree points
std::vector<test_T> query_points; //Query points
//Fill point clouds (will have a size of N*dims and M*dims)
//...
//Create KD-Tree
//Note that the data needs to be present at the CPU
auto partition_info = createKDTree<test_T, dims>(point_cloud.data(), nr_points, levels);
//CPU
std::vector<test_T> result_dists(nr_query_points*knn);
std::vector<point_i_t> result_idx(nr_query_points*knn);
KDTreeKNNSearch<test_T, test_T, dims>(partition_info,
nr_query_points, reinterpret_cast<std::array<test_T, dims>*>(query_points.data()), result_dists.data(), result_idx.data(), knn, levels);
//GPU
PartitionInfoDevice<test_T, dims>* partition_info_d = copyPartitionToGPU<test_T, dims>(partition_info);
const auto tmp = copyData<test_T, dims>(result_dists, result_idx, std::vector<std::array<test_T, dims>>(reinterpret_cast<std::array<test_T, dims>*>(query_points.data()),
reinterpret_cast<std::array<test_T, dims>*>(query_points.data()) + query_points.size() / dims));
test_T* result_dists_d = std::get<0>(tmp);
point_i_t* result_idx_d = std::get<1>(tmp);
test_T* query_points_d = std::get<2>(tmp);
KDTreeKNNGPUSearch<test_T, test_T, dims>(partition_info_d, nr_query_points,
reinterpret_cast<std::array<test_T, dims>*>(query_points_d), result_dists_d, result_idx_d, knn, levels);
auto result_gpu = copyDataBackToHost(result_dists_d, result_idx_d, nr_query_points, knn);
const auto result_dists_gpu = std::get<0>(result_gpu);
const auto result_idx_gpu = std::get<1>(result_gpu);
}Graph mode:
import numpy as np
import tensorflow as tf
from tf_nearest_neighbor import nn_distance, buildKDTree, searchKDTree
nr_refs = 1000 #Points in the KD-tree
nr_query = 100 #Points to be queried from the tree
d = 3 # Dimensions
#Create some random point clouds
points_ref = np.random.uniform(size=(nr_refs, d)).astype(np.float32) * 1e3
points_query = np.random.uniform(size=(nr_query, d)).astype(np.float32) * 1e3
#Search for the 5 nearest neighbors of each point in points_query
k = 5
points_ref_tf = tf.compat.v1.placeholder(dtype=tf.float32, shape=points_ref.shape)
points_query_tf = tf.compat.v1.placeholder(dtype=tf.float32, shape=points_query.shape)
#Build the KD-Tree in the GPU memory
build_kdtree_op = buildKDTree(points_ref_tf, levels=None) #Use maximum available levels
#Some metainformation. The important variable is part_nr which identifies the tree
structured_points, part_nr, shuffled_inds = self.sess.run(build_kdtree_op, feed_dict={points_ref_tf: points_ref})
#Search for the 5 nearest neighbors of each point in points_query
kdtree_results = searchKDTree(points_query_tf, part_nr[0], nr_nns_searches=k, shuffled_inds=shuffled_inds.astype(np.int32))
dists_knn, inds_knn = self.sess.run(kdtree_results, feed_dict={points_query_tf: points_query})or even shorter in eager mode:
import numpy as np
import tensorflow as tf
from tf_nearest_neighbor import nn_distance, buildKDTree, searchKDTree
nr_refs = 1000 #Points in the KD-tree
nr_query = 100 #Points to be queried from the tree
d = 3 # Dimensions
#Create some random point clouds
points_ref = np.random.uniform(size=(nr_refs, d)).astype(np.float32) * 1e3
points_query = np.random.uniform(size=(nr_query, d)).astype(np.float32) * 1e3
#Build the KD-Tree in the GPU memory
structured_points, part_nr, shuffled_inds = buildKDTree(points_ref, levels=None) #Use maximum available levels
#Query the KD-Tree, for the 5 nearest neighbors of each point in points_query
dists_knn, inds_knn = searchKDTree(points_query, part_nr[0], nr_nns_searches=5, shuffled_inds=shuffled_inds)Tested on Ubuntu 18.04. Exact minimal version numbers are unknown, but here are the ones with which the library was tested.
Build:
- CMake 3.13.4
- g++ 7.5.0
- Cuda 10.1
Python Interface:
- Tensorflow >= 2.0
- Numpy 1.19.2
- python 3.6
For the benchmark and tests:
- IPython 7.16.1
- Matplotlib 3.3.2
- scipy 1.5.2
Note that if you intend to install this library with an existing Tensorflow installation, you need to make sure that the CUDA and CUDNN versions match.
Inside this directory run
mkdir src/build
cd src/build
cmake .. -DCMAKE_BUILD_TYPE=RELEASE
make allCMake will fetch the library paths from the currently active python environment. This should build the library and place the library in the main directory, where it will be loaded upon importing this library.
A Dockerfile is provided that downloads the required libraries and should allow for easy testing. Docker container commands:
docker build . -t tf_nndistance_env
docker run -it --gpus all tf_nndistance_env /bin/bashInside docker:
conda activate tf_nndistance_env
cd /tf_nearest_neighbor/build
./test_kdtree && ipython ../../scripts/test_knn_unit.pyThe library contains a small C++ example that will perform a small simple test. It is run by calling ./test_kdtree in the build directory. For
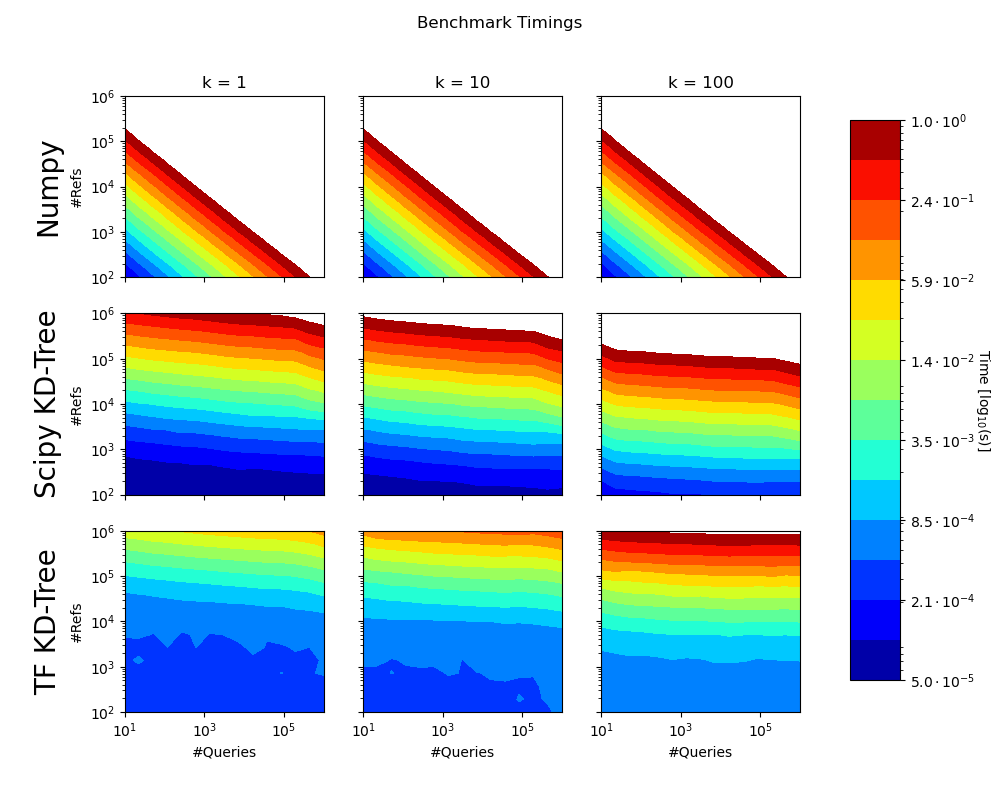
We compared both implementations to the scipy.spatial.cKDTree. Note that the benchmarks do not consider the time to build the KD-Trees, or the transfer to the GPU. Times greater than 1 second not shown.
Test Machine Specs: Intel Core i7-5820K CPU 6x @3.30GHz, 32GB of working memory and a NVidia RTX 2080 GPU.
To run the benchmark on your computer, go to the folder scripts and execute ipython benchmark.py. This will create benchmark_results.npz that can be converted to a figure using ipython plot_benchmark.py.
If this works helps you in your research, please consider acknowledging the github repository, or citing our paper from which the library originated.
@article{grandits_geasi_2021,
title = {{GEASI}: {Geodesic}-based {Earliest} {Activation} {Sites} {Identification} in cardiac models},
volume = {37},
issn = {2040-7947},
shorttitle = {{GEASI}},
url = {https://onlinelibrary.wiley.com/doi/abs/10.1002/cnm.3505},
doi = {10.1002/cnm.3505},
language = {en},
number = {8},
urldate = {2021-08-12},
journal = {International Journal for Numerical Methods in Biomedical Engineering},
author = {Grandits, Thomas and Effland, Alexander and Pock, Thomas and Krause, Rolf and Plank, Gernot and Pezzuto, Simone},
year = {2021}
}