Role of inter-related population-level host traits in determining pathogen richness and zoonotic risk
Repo for manuscript and analysis.
The preprint is on BioRxiv (preprint).
This paper is a reworking of Chapter 4 of my thesis (with commit history).
For submission to American Naturalist
It comprises simulations that test whether it is abundance, density, colony size, number of colonies or distribution area which most strongly affects pathogen richness. The simulations are run using my R package metapopEpi.
Zoonotic diseases are an increasingly important source of human infectious diseases, and reservoir host pathogen richness is a critical driver of spill-over risk. Host population-level traits such as population size and density, geographic range size and population structure have all been shown to be important determinants of host pathogen richness. However, empirically identifying the independent influences of these traits has proven difficult as many of these traits directly depend on each other. Here we develop a mechanistic, metapopulation, susceptible-infected-recovered model to identify the influences of independent population-level traits on the ability of a newly evolved pathogen to invade and persist in host populations in the presence of an endemic pathogen, using a case study of bats; a highly social mammalian order. We show that larger population and group sizes had a greater influence on the chances of pathogen invasion and persistence than increased population densities (and therefore decreased population structure) and number of groups. As anthropogenic change affects these traits to different extents, this increased understanding of how traits independently determine pathogen richness will aid in predicting future zoonotic spill-over risk.
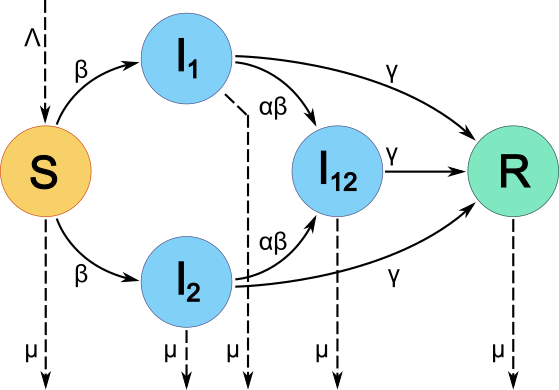
Figure 1: Schematic of the two-pathogen SIR model used. Individuals are in one of five epidemiological classes, susceptible (orange, S), infected with Pathogen 1, Pathogen 2 or both (blue, I1, I2, I12, respectively) or recovered and immune from further infection (green, R). Transitions between classes occur as indicated by solid arrows and depend on transmission rate (β), coinfection adjustment factor (α) and recovery rate (γ). Births (Λ) and deaths (μ) are indicated by dashed arrows. Note that individuals in I12 move into R, not back to I1 or I2. That is, recovery from one pathogen causes immediate recovery from the other pathogen.
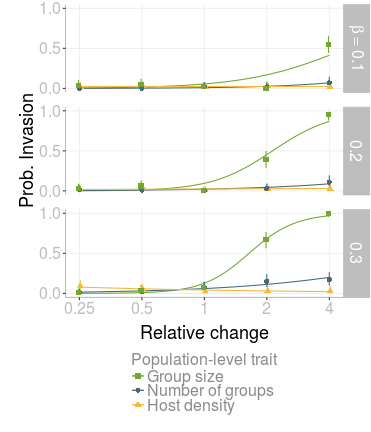
Figure 2: Comparison of the effect of population-level factors on probability of invasion. Population-level factors are group size (green lines, squares), number of groups (blue lines, circles) and host density (yellow lines, triangles). The x-axis shows the change (x0.25, 0.5, 1, 2 and 4) in each of these factors relative to the default value. Default values are: number of groups = 20, group size = 400 and host density = 0.8 animals per km2. Each point is the mean of 100 simulations and bars are 95% confidence intervals. Curves are binomial GLM regression fits. Relationships are shown separately for each transmission value, β.