- Neuron Coverage (NC),
- Cell Coverage (CC),
- Gate Coverage (GC),
- Sequence Coverage (SQ)
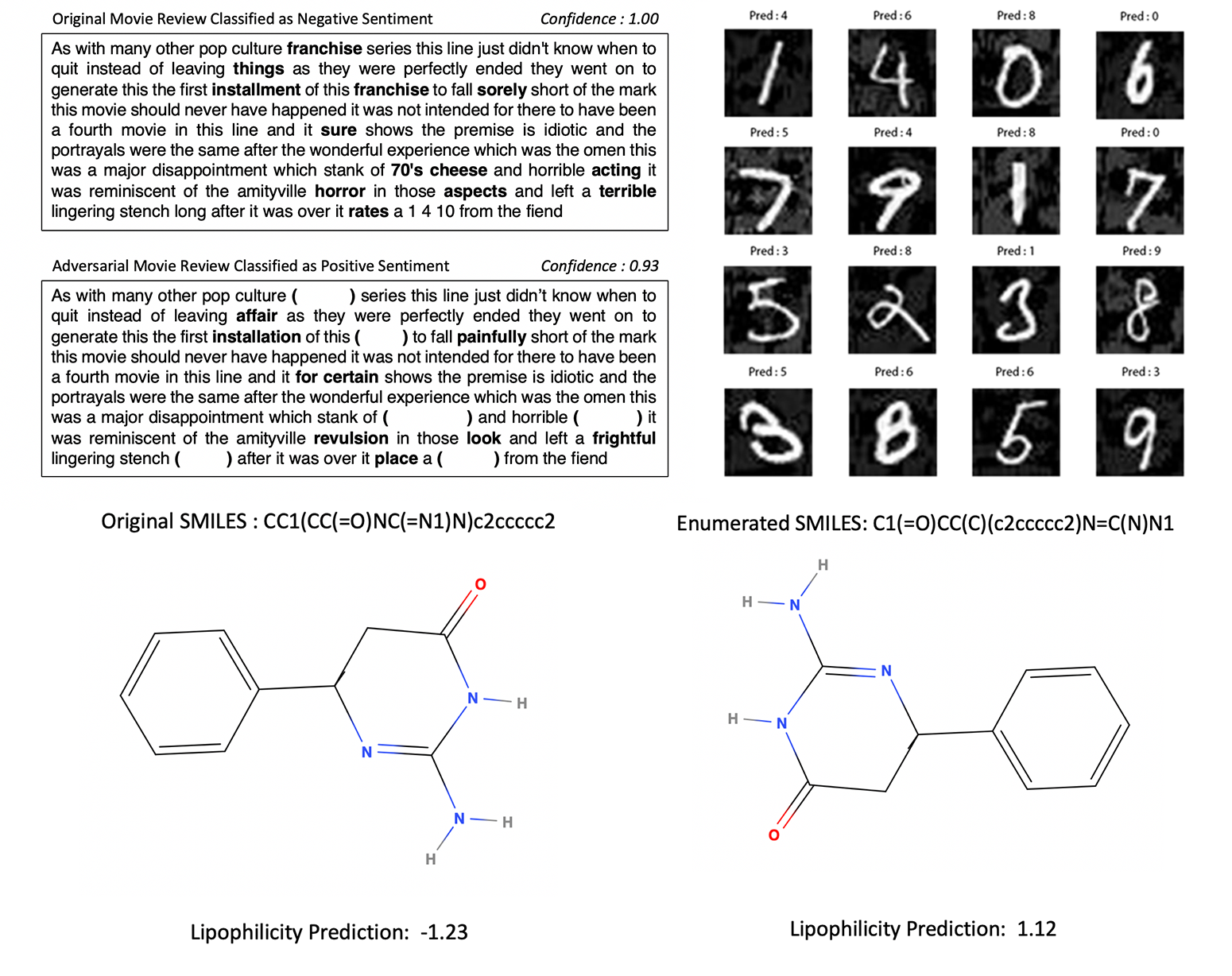
- Sentiment Analysis,
- MNIST Handwritten Digits,
- Lipophilicity Prediction (Physical Chemistry)
-
rdkit (https://www.rdkit.org/docs/Install.html), by running the following commands:
conda create -c rdkit -n my-rdkit-env rdkit conda activate my-rdkit-env
Note: with the above commands, we create a new virtual environment dedicated for rdkit. Below, every time one needs to run the program, he/she needs to activate the my-rdkit-env
-
Other packages including
conda install -c menpo opencv keras nltk matplotlib pip install saxpy sklearn
We have two commands to run testing procedure and to run result analysis procedure, respectively.
python main.py --model <modelName>
--TestCaseNum <Num. of Test Cases>
--TargMetri <Terminate Metrics>
--CoverageStop <Terminate Coverage Rate>
--threshold_CC <CC threshold>
--threshold_GC <GC threshold>
--symbols_SQ <Num. of symbols>
--seq <seq in cells to test>
[--mode <modeName>]
--minimalTest <if generate minimal test set>
--output <output file path>
where
- <modelName> can be in {sentiment,mnist,lipo}
- <Num. of Test Cases> is expected number of test cases
- <Termination Metrics> can be in {CC,GC,SQN,SQP}
- <Termination Coverage Rate> is expected coverage rate of Termination Metric
- <CC threshold> can be in [3,9]
- <GC threshold> can be in [0,1]
- <Num. of symbols> can be in {2,3}
- <seq in cells to test> can be in {mnist:[19,23],sentiment:[480:484],lipo:[70,74]}
- <modeName> can be in {train,test} with default value test
- <Generate minimal test set> can be in {0: No, 1: Yes}
- <output file path> specifies the path to the output file
For example, we can run the following
python main.py --model mnist --TestCaseNum 2000 --TargMetri CC --CoverageStop 0.9 --threshold_CC 6 --threshold_GC 0.8 --symbols_SQ 2 --seq [19,23] --minimalTest 0 --output log_folder/record.txt
which says that, we are working with MNIST model, and the test case generation will terminate when either the number of test cases is over 2000 or the target metric, Cell Coverage, reaches 0.9 coverage rate. We need to specify other parameters including threshold_CC, threshold_GC, symbols_SQ, seq. Moreover, we do not ask for the generation of miminal test suite, and the log is generated to the file log_folder/record.txt.
readfile.py can read the log file log_folder\record.txt and .MAT file of test conditions counting. Several figures includes the coverage updating information for all metrics and test conditions statistics plot are generated.
python readfile.py --output log_folder/record.txt --metrcis all