
(c) 2022-2024 Timothy James Becker
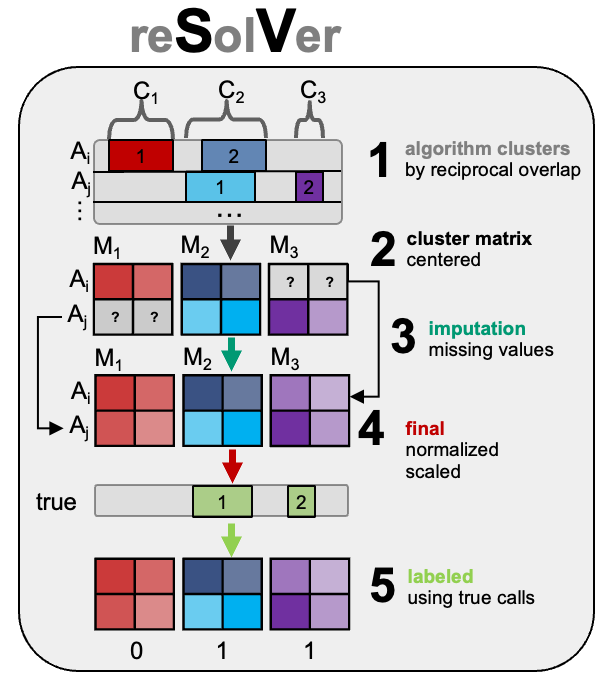
Resolving multiple structural variation callers and platforms by matrix transformation and imputation
3.11+ python
1.26+ numpy
0.22+ pysam
1.2+ scikit-learn
pip is not setup at this time, simply clone the repo and then use the resolver.py script to run model training, testing and prediciton as described below in the usage section.
resolver.py \
--in_dirs dir1,dir2,dir3... \
--pred_dirs dirP \
--out_dir dirP_result \
--seqs chr1,chr2,... \
--split 0.5
--iterations 30 \
--sv_types DEL,DUP,INV,INS \
--cpus 12--in_dirs: comma seperated list of stage-mapped SV sample folders. Each sample folder should contain SV calls that have simplified naming conventions: samplename_SID. For example sample HG00096 would have the Lumpy SV file named according to the stage ID as HG00096_S18.vcf. True calls that are given from 1000 Genomes or other projects act as supervision for the machine learning have the unique stage ID as 9. For example HG00096_S0.vcf is the true set of SV calls for sample HG00096.
--pred_dir: comma seperated directors to run prediction on. This would be where you would put new calls that you wanted to run reSolVer models on that were pretrained such as the resolver_g1kp3 or resolver_multi models.
--out_dir: output directory where the chrod,clust,models,train,vcam,vcfs folders and files will be generated.
--seqs: sequences that the training,testing,and validation (or prediction) should be perfromed on. From standard VCF calling you would want all the reference chromosomes of interest.
--split: used for training to set the amount of data that will be given to the modeling during the fitting or training phase. The remaining proportion will then be split into the testing phase to estimate the f1 accuracy and then the final amount which are validation are never seen by the model or estimated. They can be used to produce an unbiased final model estimate during the prediction once the models parameters are fixed or to calibrate the probabilty performance.
--iterations: the number of iterations(random tries) that the modeling will use to fit during training. Higher iterations will often overfit and low will underfit to the training portion of the data.
--sv_types: comma seperated list of SV types, can be used to build models for every type or just for one at a time in case of || training.
--cpus: the number of computer cores to use for data processing and model fiting
