This repository holds the code for RefineGAN,
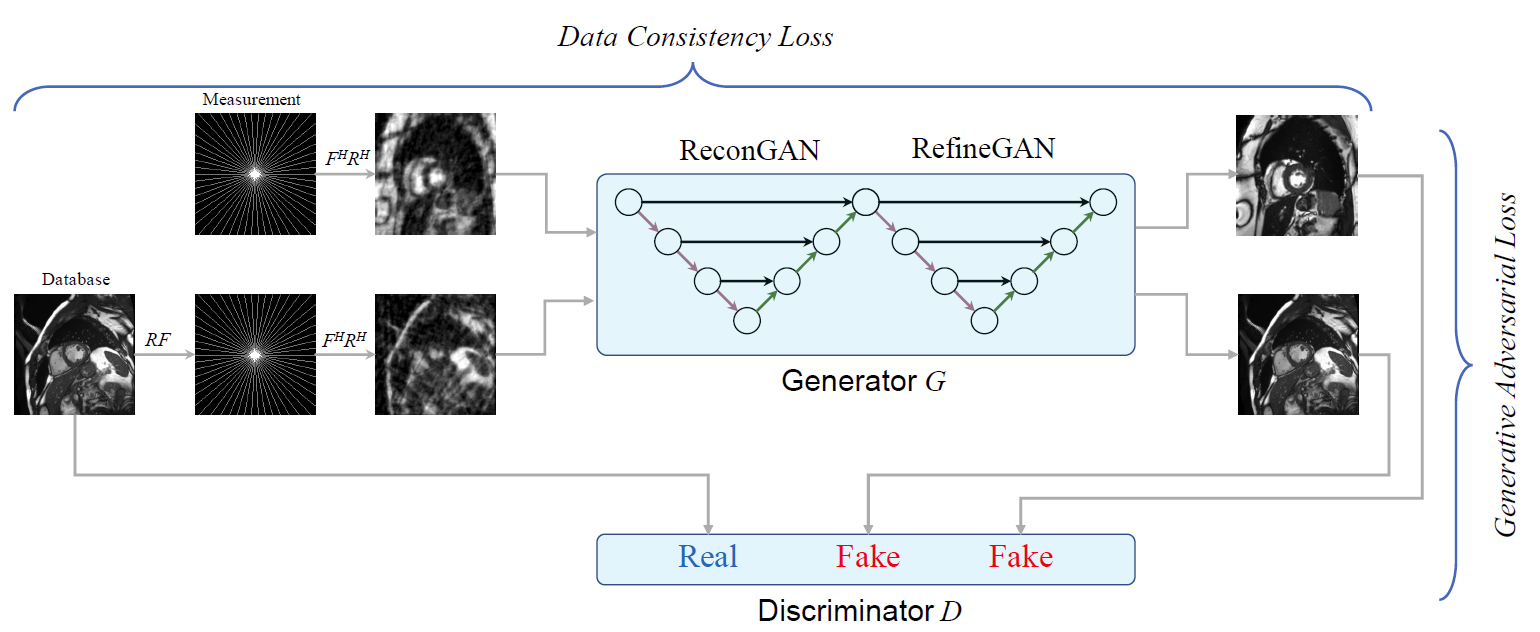 Overview of the proposed method: it aims to reconstruct the images which are satisfied the constraint of under-sampled measurement data; and
whether those look similar to the fully aliasing-free results. Additionally, if the fully sampled images taken from the database go through the same process of
under-sampling acceleration; we can still receive the reconstruction as expected to the original images.
Overview of the proposed method: it aims to reconstruct the images which are satisfied the constraint of under-sampled measurement data; and
whether those look similar to the fully aliasing-free results. Additionally, if the fully sampled images taken from the database go through the same process of
under-sampling acceleration; we can still receive the reconstruction as expected to the original images.
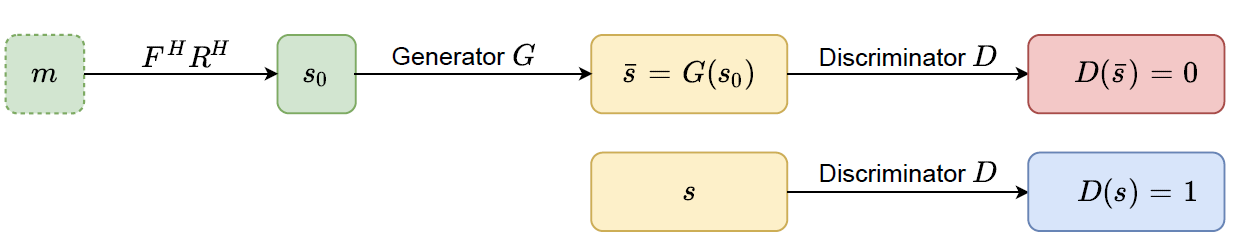 Two learning processes are trained adversarially to achieve better reconstruction from generator G and to fool the ability of recognizing the real or
fake MR image from discriminator D
Two learning processes are trained adversarially to achieve better reconstruction from generator G and to fool the ability of recognizing the real or
fake MR image from discriminator D
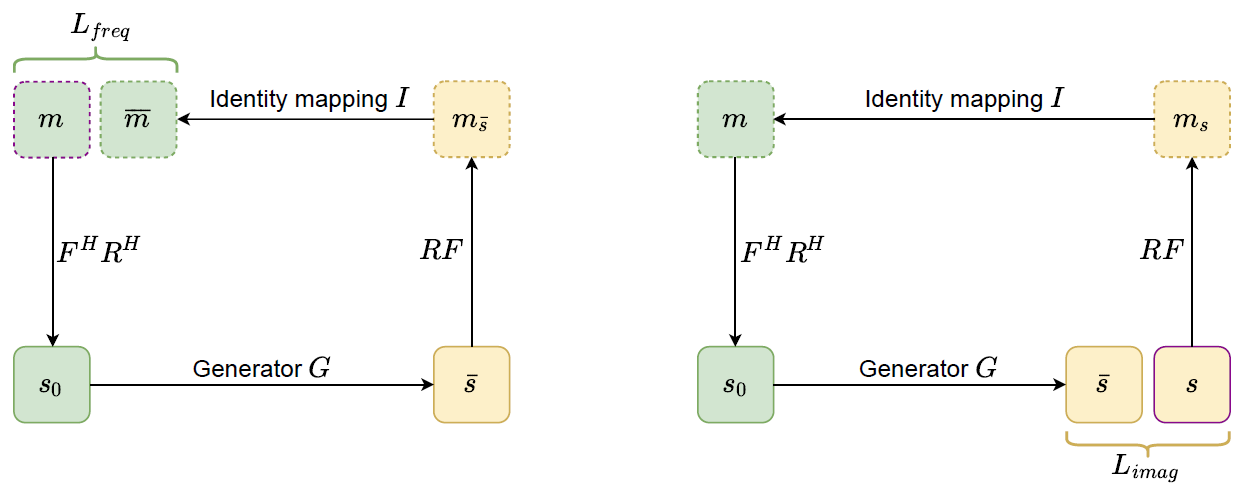 The cyclic data consistency loss, which is a combination of under-sampled frequency loss and the fully reconstructed image loss.
The cyclic data consistency loss, which is a combination of under-sampled frequency loss and the fully reconstructed image loss.
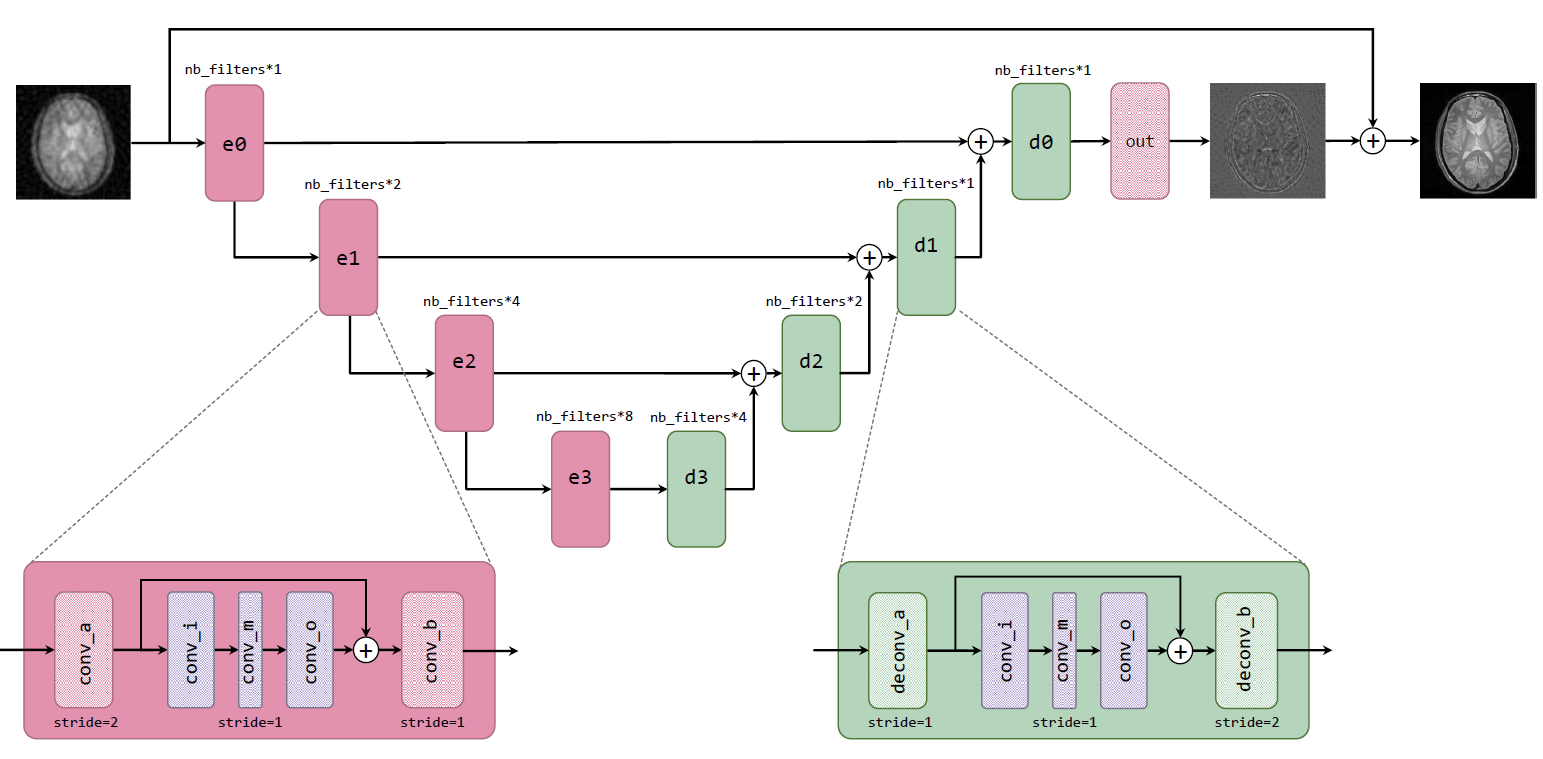 Generator G, built by basic building blocks, can reconstruct inverse amplitude of the residual component causes by reconstruction from under-sampled
k-space data. The final result is obtained by adding the zero-filling reconstruction to the output of G
Generator G, built by basic building blocks, can reconstruct inverse amplitude of the residual component causes by reconstruction from under-sampled
k-space data. The final result is obtained by adding the zero-filling reconstruction to the output of G
It is developed for research purposes only and not for commercialization. If you use it, please refer to our work.
@ARTICLE{8327637,
author={T. M. Quan and T. Nguyen-Duc and W. Jeong},
journal={IEEE Transactions on Medical Imaging},
title={Compressed Sensing MRI Reconstruction Using a Generative Adversarial Network With a Cyclic Loss},
year={2018},
volume={37},
number={6},
pages={1488-1497},
doi={10.1109/TMI.2018.2820120},
ISSN={0278-0062},
month={June},
}
Directory structure of data:
tree data
data/
├── brain
│ ├── db_train
│ └── db_valid
├── knees
│ ├── db_train
│ └── db_valid
└── mask
├── cartes
│ ├── mask_1
│ ├── mask_2
│ ├── ...
│ └── mask_9
├── gauss
│ ├── mask_1
│ ├── mask_2
│ ├── ...
│ └── mask_9
├── radial
│ ├── mask_1
│ ├── mask_2
│ ├── ...
│ └── mask_9
└── spiral
├── mask_1
├── mask_2
├── ...
└── mask_9
Brain data is used for magnitude-value experiment, it is extracted from http://brain-development.org/ixi-dataset/
Knees data is used for complex-value experiment, it is extracted from http://mridata.org
Prerequisites
sudo pip install tensorflow_gpu==1.4.0
sudo pip install tensorpack==0.8.2
sudo pip install scikit-image==0.13.0
sudo pip install whatever-missing_libraries
To begin, the template for such an experiment is provided in exp_dset_RefineGAN_mask_strategy_rate.py
For example, if you want to run the training and testing for case knees data, mask radial 10%, please make a soft link to the experiment name, like this
ln -s exp_dset_RefineGAN_mask_strategy_rate.py \
exp_knees_RefineGAN_mask_radial_1.py
To train the model
python exp_knees_RefineGAN_mask_radial_1.py \
--gpu='0' \
--imageDir='data/knees/db_train/' \
--labelDir='data/knees/db_train/' \
--maskDir='data/mask/radial/mask_1/'
Checkpoint of training will be save to directory train_log
To test the model
mkdir result
python exp_knees_RefineGAN_mask_radial_1.py \
--gpu='0' \
--imageDir='data/knees/db_valid/' \
--labelDir='data/knees/db_valid/' \
--maskDir='data/mask/radial/mask_1/' \
--sample='result/exp_knees_RefineGAN_mask_radial_1/' \
--load='train_log/exp_knees_RefineGAN_mask_radial_1/max-validation_PSNR_boost_A.data-00000-of-00001'
The authors would like to thank Dr. Yoonho Nam for the helpful discussion and MRI data, and Yuxin Wu for the help on Tensorpack.