outbreakR provides simple tools to extract corrected indices and insect outbreak reconstruction data from dendrochronological time series data.
Install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("toddellis/outbreakR")outbreakR comes with two demo datasets:
ob_host: A host dataset comprised of tree-level ring-width indices from Douglas-fir trees sampled across four study sites in 2014.ob_nonhost: A nonhost master chronology – the first principal component – built from ponderosa pine trees sampled across study sites from the same region.
library(outbreakR)
head(ob_host)
#> # A tibble: 6 x 69
#> year MPD01 MPD02 MPD03 MPD04 MPD05 MPD06 MPD07 MPD08
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1685 0.696 NA NA NA NA NA NA NA
#> 2 1686 0.690 NA NA NA NA NA NA NA
#> 3 1687 1.09 NA NA NA NA NA NA NA
#> 4 1688 0.844 NA NA NA NA 0.547 NA NA
#> 5 1689 1.15 NA NA NA NA 0.813 NA NA
#> 6 1690 0.695 NA NA NA NA 0.264 NA NA
#> # ... with 60 more variables: MPD09 <dbl>, MPD10 <dbl>,
#> # MPD11 <dbl>, MPD12 <dbl>, MPD14 <dbl>, MPD15 <dbl>,
#> # MPD16 <dbl>, SMD01 <dbl>, SMD02 <dbl>, SMD03 <dbl>,
#> # SMD04 <dbl>, SMD05 <dbl>, SMD06 <dbl>, SMD07 <dbl>,
#> # SMD08 <dbl>, SMD09 <dbl>, SMD10 <dbl>, SMD11 <dbl>,
#> # SMD12 <dbl>, SMD13 <dbl>, SMD14 <dbl>, SMD15 <dbl>,
#> # SMD16 <dbl>, SMD17 <dbl>, SMD18 <dbl>, TMD01 <dbl>,
#> # TMD02 <dbl>, TMD03 <dbl>, TMD04 <dbl>, TMD05 <dbl>,
#> # TMD06 <dbl>, TMD07 <dbl>, TMD08 <dbl>, TMD09 <dbl>,
#> # TMD10 <dbl>, TMD11 <dbl>, TMD12 <dbl>, TMD13 <dbl>,
#> # TMD14 <dbl>, TMD15 <dbl>, TMD16 <dbl>, TMD17 <dbl>,
#> # TMD18 <dbl>, TMD19 <dbl>, TMD20 <dbl>, VLD01 <dbl>,
#> # VLD02 <dbl>, VLD03 <dbl>, VLD04 <dbl>, VLD05 <dbl>,
#> # VLD06 <dbl>, VLD07 <dbl>, VLD08 <dbl>, VLD09 <dbl>,
#> # VLD10 <dbl>, VLD11 <dbl>, VLD12 <dbl>, VLD13 <dbl>,
#> # VLD14 <dbl>, VLD15 <dbl>
head(ob_nonhost)
#> # A tibble: 6 x 2
#> year nonhost
#> <dbl> <dbl>
#> 1 1685 -0.106
#> 2 1686 0.183
#> 3 1687 -0.657
#> 4 1688 -1.97
#> 5 1689 -0.360
#> 6 1690 -1.45To extract insect outbreak data, outbreakR uses two simple functions.
The first produces corrected indices by removing climatic noise from
the host tree-level data, leaving an assumed biological signal.
The default settings assume wide-format data, as expected from the
output of dplR’s RWI functions.
Just be aware of the additional settings for cor_index().
foo <- cor_index(ob_host, ob_nonhost)
head(foo)
#> # A tibble: 6 x 5
#> # Groups: tree_id [1]
#> year tree_id host nonhost ci
#> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 1685 MPD01 0.696 -0.106 -1.05
#> 2 1686 MPD01 0.690 0.183 -1.25
#> 3 1687 MPD01 1.09 -0.657 0.782
#> 4 1688 MPD01 0.844 -1.97 0.678
#> 5 1689 MPD01 1.15 -0.360 0.818
#> 6 1690 MPD01 0.695 -1.45 -0.203As an alternative, it also takes long-format data:
ob_host %>%
tidyr::gather('id', 'rwi', -year) %>%
cor_index(.,
ob_nonhost,
format = 'long') %>%
head()
#> # A tibble: 6 x 5
#> # Groups: tree_id [1]
#> year tree_id host nonhost ci
#> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 1685 MPD01 0.696 -0.106 -1.05
#> 2 1686 MPD01 0.690 0.183 -1.25
#> 3 1687 MPD01 1.09 -0.657 0.782
#> 4 1688 MPD01 0.844 -1.97 0.678
#> 5 1689 MPD01 1.15 -0.360 0.818
#> 6 1690 MPD01 0.695 -1.45 -0.203Many dendrochronological datasets come in wide format rather than long, and treat years as the row names. This can also be handled here, but it’s really recommended that you avoid deprecated data formats like this:
## Reformat both host and nonhost to mimic older formatting
host_old <- ob_host %>%
as.data.frame()
row.names(host_old) <- host_old$year
host_old <- host_old %>%
dplyr::select(-year)
nonhost_old <- ob_nonhost %>%
as.data.frame()
row.names(nonhost_old) <- nonhost_old$year
nonhost_old <- nonhost_old %>%
dplyr::select(-year)
head(host_old)
#> MPD01 MPD02 MPD03 MPD04 MPD05 MPD06 MPD07
#> 1685 0.6960065 NA NA NA NA NA NA
#> 1686 0.6897567 NA NA NA NA NA NA
#> 1687 1.0949695 NA NA NA NA NA NA
#> 1688 0.8440234 NA NA NA NA 0.5471116 NA
#> 1689 1.1547135 NA NA NA NA 0.8128844 NA
#> 1690 0.6954994 NA NA NA NA 0.2642121 NA
#> MPD08 MPD09 MPD10 MPD11 MPD12 MPD14 MPD15
#> 1685 NA NA NA NA NA NA 1.6632179
#> 1686 NA NA NA NA NA NA 0.5644853
#> 1687 NA NA NA NA NA NA 0.5698452
#> 1688 NA NA NA NA NA NA 0.5048660
#> 1689 NA NA NA NA NA NA 0.4808650
#> 1690 NA NA NA NA NA NA 0.3278609
#> MPD16 SMD01 SMD02 SMD03 SMD04 SMD05 SMD06 SMD07
#> 1685 NA NA NA NA NA NA NA NA
#> 1686 NA NA NA NA NA NA NA NA
#> 1687 NA NA NA NA NA NA NA NA
#> 1688 NA NA NA NA NA NA NA NA
#> 1689 NA NA NA NA NA NA NA NA
#> 1690 NA NA NA NA NA NA NA NA
#> SMD08 SMD09 SMD10 SMD11 SMD12 SMD13 SMD14 SMD15
#> 1685 NA NA NA NA NA NA NA NA
#> 1686 NA NA NA NA NA NA NA NA
#> 1687 NA NA NA NA NA NA NA NA
#> 1688 NA NA NA NA NA NA NA NA
#> 1689 NA NA NA NA NA NA NA NA
#> 1690 NA NA NA NA NA NA NA NA
#> SMD16 SMD17 SMD18 TMD01 TMD02 TMD03 TMD04
#> 1685 NA NA NA 0.3331848 NA NA 0.9105027
#> 1686 NA NA NA 0.3211430 NA NA 0.7801712
#> 1687 NA NA NA 0.1771122 NA NA 0.8838990
#> 1688 NA NA NA 0.2139429 NA NA 0.7627637
#> 1689 NA NA NA 0.5547610 NA NA 0.9994772
#> 1690 NA NA NA 0.3930280 NA NA 0.7515191
#> TMD05 TMD06 TMD07 TMD08 TMD09 TMD10 TMD11
#> 1685 NA NA NA NA NA NA NA
#> 1686 NA NA NA NA NA NA NA
#> 1687 NA NA NA NA NA NA NA
#> 1688 NA NA NA NA NA NA NA
#> 1689 NA NA NA NA NA NA NA
#> 1690 NA NA NA NA NA NA NA
#> TMD12 TMD13 TMD14 TMD15 TMD16 TMD17
#> 1685 0.8555250 0.8575240 NA NA 0.8227725 NA
#> 1686 0.6882582 0.7379745 NA NA 0.9142322 NA
#> 1687 0.7302570 0.8357733 NA NA 0.5854386 NA
#> 1688 0.4114042 0.4655927 NA NA 0.3042688 NA
#> 1689 0.6241529 0.7525962 NA NA 0.8982428 NA
#> 1690 0.4646850 0.6197168 NA NA 0.7747778 NA
#> TMD18 TMD19 TMD20 VLD01 VLD02 VLD03 VLD04 VLD05
#> 1685 NA NA NA NA NA NA NA NA
#> 1686 NA NA NA NA NA NA NA NA
#> 1687 NA NA NA NA NA NA NA NA
#> 1688 NA NA NA NA NA NA NA NA
#> 1689 NA NA NA NA NA NA NA NA
#> 1690 NA NA NA NA NA NA NA NA
#> VLD06 VLD07 VLD08 VLD09 VLD10 VLD11 VLD12 VLD13
#> 1685 NA NA NA NA NA NA NA NA
#> 1686 NA NA NA NA NA NA NA NA
#> 1687 NA NA NA NA NA NA NA NA
#> 1688 NA NA NA NA NA NA NA NA
#> 1689 NA NA NA NA NA NA NA NA
#> 1690 NA NA NA NA NA NA NA NA
#> VLD14 VLD15
#> 1685 NA NA
#> 1686 NA NA
#> 1687 NA NA
#> 1688 NA NA
#> 1689 NA NA
#> 1690 NA NA
## Specify years_as_rownames = TRUE
## Note: This does not work with long-format data
cor_index(host_old, nonhost_old, years_as_rownames = T) %>%
head()
#> [1] "Please avoid using years as rownames."
#> # A tibble: 6 x 5
#> # Groups: tree_id [1]
#> year tree_id host nonhost ci
#> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 1685 MPD01 0.696 -0.106 -1.05
#> 2 1686 MPD01 0.690 0.183 -1.25
#> 3 1687 MPD01 1.09 -0.657 0.782
#> 4 1688 MPD01 0.844 -1.97 0.678
#> 5 1689 MPD01 1.15 -0.360 0.818
#> 6 1690 MPD01 0.695 -1.45 -0.203
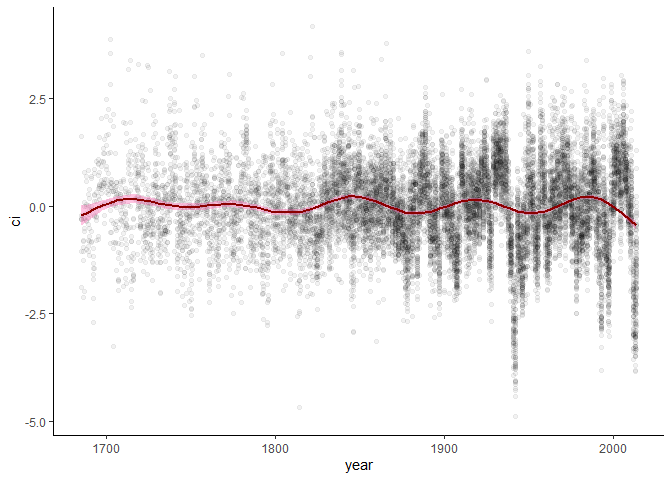
foo %>%
ggplot(aes(x = year, y = ci)) +
theme_classic() +
geom_point(alpha = 0.05) +
geom_smooth(method = 'gam',
formula = y ~ s(x, k = 12),
color = 'darkred',
fill = 'hotpink')The outbreak function extracts either binary outbreak/non-outbreak
time series data at the tree level, or produces proportional outbreaks
showing the percentage of trees with an outbreak in a given year.
outbreak(foo) %>%
tail(10)
#> # A tibble: 10 x 2
#> year outbreakProp
#> <dbl> <dbl>
#> 1 2005 2.99
#> 2 2006 2.99
#> 3 2007 2.99
#> 4 2008 5.97
#> 5 2009 40.3
#> 6 2010 65.7
#> 7 2011 67.2
#> 8 2012 68.2
#> 9 2013 68.2
#> 10 2014 64.6
outbreak(foo, prop = FALSE) %>%
tail(10)
#> # A tibble: 10 x 6
#> year tree_id host nonhost ci outbreak
#> <dbl> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 2005 VLD15 0.699 -0.624 -0.494 1
#> 2 2006 VLD15 0.864 -2.29 1.18 1
#> 3 2007 VLD15 0.427 -2.53 -0.0447 1
#> 4 2008 VLD15 0.617 -1.95 0.161 1
#> 5 2009 VLD15 0.652 -1.23 -0.224 1
#> 6 2010 VLD15 0.858 0.626 -0.854 1
#> 7 2011 VLD15 1.01 0.480 -0.276 1
#> 8 2012 VLD15 0.409 2.14 -3.32 1
#> 9 2013 VLD15 0.482 2.52 -3.35 1
#> 10 2014 VLD15 0.972 1.77 -1.28 1
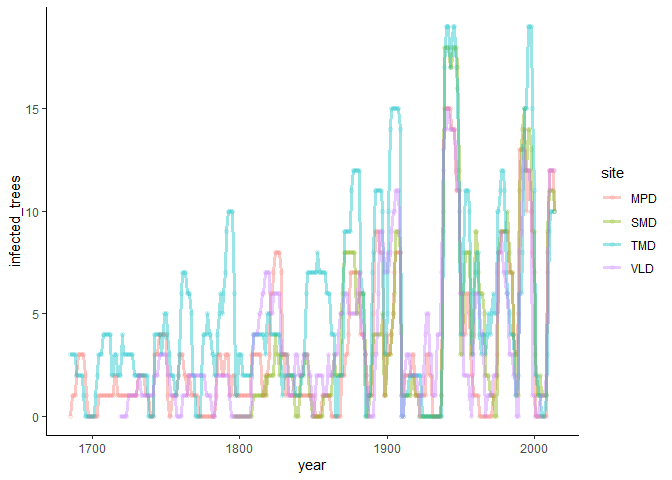
outbreak(foo, prop = FALSE) %>%
mutate(site = substr(tree_id, 1, 3)) %>%
group_by(site, year) %>%
summarise(infected_trees = sum(outbreak)) %>%
ggplot(aes(x = year, y = infected_trees, color = site)) +
theme_classic() +
geom_point(alpha = 0.1) +
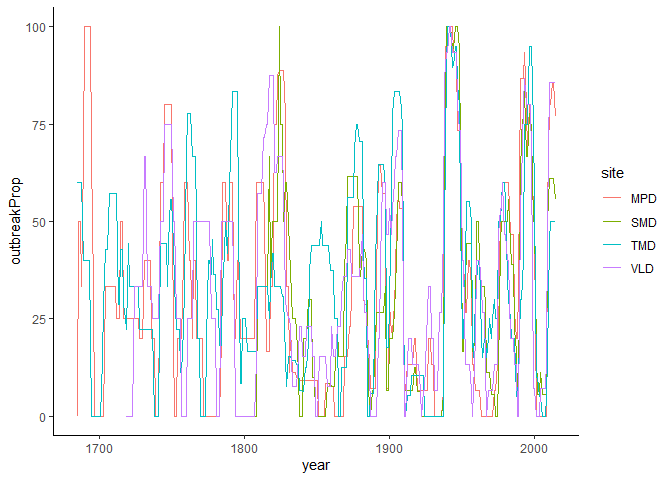
geom_line(size = 1.2, alpha = 0.4)We can extract the sites here and run it individually per site. I may try to implement this in the future, but for now, this would provide a workaround:
## add site variable
foo <- foo %>%
dplyr::mutate(site = substr(tree_id, 1, 3))
## pull out unique sites
sites <- foo %>%
.$site %>%
unique()
## create empty list to fill
site_outbreaks <- list()
## loop through site list
for (i in 1:length(sites)) {
site_outbreaks[[i]] <- foo %>%
dplyr::filter(site == sites[i]) %>%
outbreak() %>%
dplyr::mutate(site = sites[i])
}
site_outbreaks <- dplyr::bind_rows(site_outbreaks)
site_outbreaks %>%
ggplot(aes(x = year, y = outbreakProp, color = site)) +
geom_line() +
theme_classic()Quite hideous at this stage, but will be fixed a bit in the next update.