specieshindex is a package that aims to gauge scientific influence of
different species mainly using the h-index.
To get this package to work, make sure you have the following packages installed.
#Installation from GitHub
install.packages("rscopus")
install.packages("wosr")
install.packages("rbace")
install.packages("taxize")
install.packages("XML")
install.packages("jsonlite")
install.packages("httr")
install.packages("dplyr")
install.packages("data.table")
install.packages("tidyr")
devtools::install_github("jessicatytam/specieshindex", force = TRUE, build_vignettes = FALSE)
#Load the library
library(specieshindex)You can find the vignette here for more detailed instructions and the full list of functions here.
The Count and Fetch functions allow the addition of keywords using Boolean operators to restrict the domain of the search. Although you can simply use keywords such as “conservation”, you will find that using “conserv*” will yield more results. The “*” (or wildcard) used here searches for any words with the prefix “conserv”, e.g. conservation, conserve, conservatory, etc. Find out more about search language here and here.
Some species have had their classification changed in the past, resulting in multiple binomial names and synonyms. Synonyms can be added to the search strings to get the maximum hits. If you have more than 1 synonym, you can parse a list (the list should be named “synonyms”) into the argument.
Make sure you are connected to the internet via institutional access or acquire a VPN from your institution if you are working from home. Alternatively, if you are a subscriber of Scopus already, you can ignore this step.
To connect and download citation information from Scopus legally, you will absolutely need an API key. Here are the steps to obtain the key.
- Go to https://dev.elsevier.com/ and click on the button
I want an API key. - Create an account and log in.
- Go to the
My API Keytab on top of the page and clickCreate API Key. - Read the legal documents and check the boxes.
You are required to be at your institution for this to work since the API is accessed via the IP address.
It is recommended that you have your IP address whitelisted. You can do it here.
Here is a quick demonstration of how the package works.
You will need to set up your API key / session ID before gaining access to the databases. Run the following lines of code to do so.
#Scopus
apikey <- "your_scopus_api_key"
#Web of Science
sid <- auth(username = NULL, password = NULL)You won’t have to set this again until your next session. You are required to be at your institution for this to work since the API is accessed via the IP address.
Multiple databases have been incorporated into specieshindex,
including Scopus, Web of Science, and Lens. To differentiate between
them, the suffix of the Count and Fetch functions have been labeled with
the database’s name, with the exception of Scopus.
#Scopus requests
CountSpT(db = "scopus", genus = "Bettongia", species = "penicillata")
FetchSpT(db = "scopus", genus = "Bettongia", species = "penicillata")
CountGenusT(db = "scopus", genus = "Bettongia")
FetchGenusT(db = "scopus", genus = "Bettongia")
#Web of science requests
#No tokens or api keys needed if session ID has been set as shown previously
CountSpT(db = "wos", genus = "Bettongia", species = "penicillata")
FetchSpT(db = "wos", genus = "Bettongia", species = "penicillata")
CountGenusT(db = "wos", genus = "Bettongia")
FetchGenusT(db = "wos", genus = "Bettongia")
#BASE requests
#Fetch functions are not available for BASE
CountSpT(db = "base", genus = "Bettongia", species = "penicillata")
CountGenusT(db = "base", genus = "Bettongia")If you are only interested in knowing how many publications there are on
Scopus, you can run the Count functions. Use CountSpT() for title only
or CountSpTAK() for title+abstract+keywords.
#Count citation data
CountSpT("scopus", "Bettongia", "penicillata")
CountSpTAK("scopus", "Bettongia", "penicillata")
#Example including additional keywords
CountSpTAK("scopus", "Phascolarctos", "cinereus",
additionalkeywords = "(consrv* OR protect* OR reintrod* OR restor*)")
#search string: TITLE-ABS-KEY("Phascolarctos cinereus" AND (consrv* OR protect* OR reintrod* OR restor*))
#Example including synonyms
CountSpT("scopus", "Osphranter", "rufus",
synonyms = "Macropus rufus", additionalkeywords = "conserv*")
#search string: TITLE(("Osphranter rufus" OR "Macropus rufus") AND conserv*)In order to calculate the indices, you will need to download the
citation records. The parameters of the Count and Fetch functions are
exactly the same. Let’s say you want to compare the species h-index of a
few marsupials. First, you would need to download the citation
information using either FetchSpT() for title only or FetchSpTAK()
for title+abstract+keywords. Remember to use binomial names.
#Extract citation data
Woylie <- FetchSpTAK("scopus", "Bettongia", "penicillata")
Quokka <- FetchSpTAK("scopus", "Setonix", "brachyurus")
Platypus <- FetchSpTAK("scopus", "Ornithorhynchus", "anatinus")
Koala <- FetchSpTAK("scopus", "Phascolarctos", "cinereus")Now that you have the data, you can use the Allindices() function to
create a dataframe that shows their indices.
# Calculate indices
W <- Allindices(Woylie, genus = "Bettongia", species = "penicillata")
Q <- Allindices(Quokka, genus = "Setonix", species = "brachyurus")
P <- Allindices(Platypus, genus = "Ornithorhynchus", species = "anatinus")
K <- Allindices(Koala, genus = "Phascolarctos", species = "cinereus")
CombineSp <- dplyr::bind_rows(W, Q, P, K) #combining the citation records
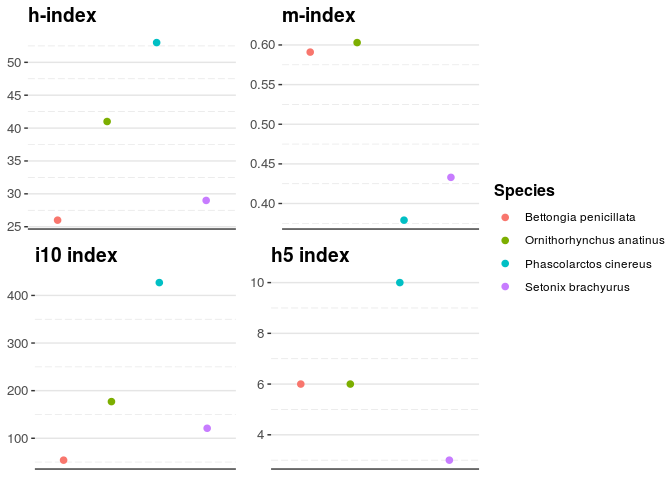
CombineSp## genus_species species genus publications citations
## 1 Bettongia penicillata penicillata Bettongia 113 1903
## 2 Setonix brachyurus brachyurus Setonix 242 3427
## 3 Ornithorhynchus anatinus anatinus Ornithorhynchus 321 6365
## 4 Phascolarctos cinereus cinereus Phascolarctos 773 14291
## journals years_publishing h m i10 h5
## 1 55 44 26 0.591 54 6
## 2 107 67 29 0.433 121 3
## 3 153 68 41 0.603 177 6
## 4 227 140 53 0.379 427 10
Once you are happy with your dataset, you can make some nice plots.
Using plotAllindices(), we can compare the indices against each other.
plotAllindices(CombineSp)Figure 1. The h-index, m-index, i10 index, and h5 index of the Woylie, Platypus, Koala, and Quokka.
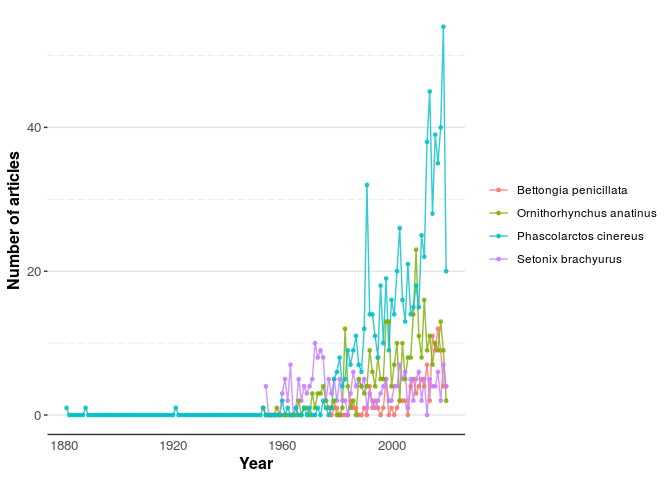
You can also visualise the total publication per year with getYear()
and plotPub().
extract_year_W <- getYear(data = Woylie, genus = "Bettongia", species = "penicillata")
extract_year_Q <- getYear(data = Quokka, genus = "Setonix", species = "brachyurus")
extract_year_P <- getYear(data = Platypus, genus = "Ornithorhynchus", species = "anatinus")
extract_year_K <- getYear(data = Koala, genus = "Phascolarctos", species = "cinereus")
Combine_pub <- rbind(extract_year_W, extract_year_Q, extract_year_P, extract_year_K)
plotPub(Combine_pub)Figure 2. The total number of publications per year of the Woylie, Platypus, Koala, and Quokka.
specieshindex is enabled by Scopus, Web of Science, and BASE.
To propose any bug fixes or new features, please refer to our community guidelines.