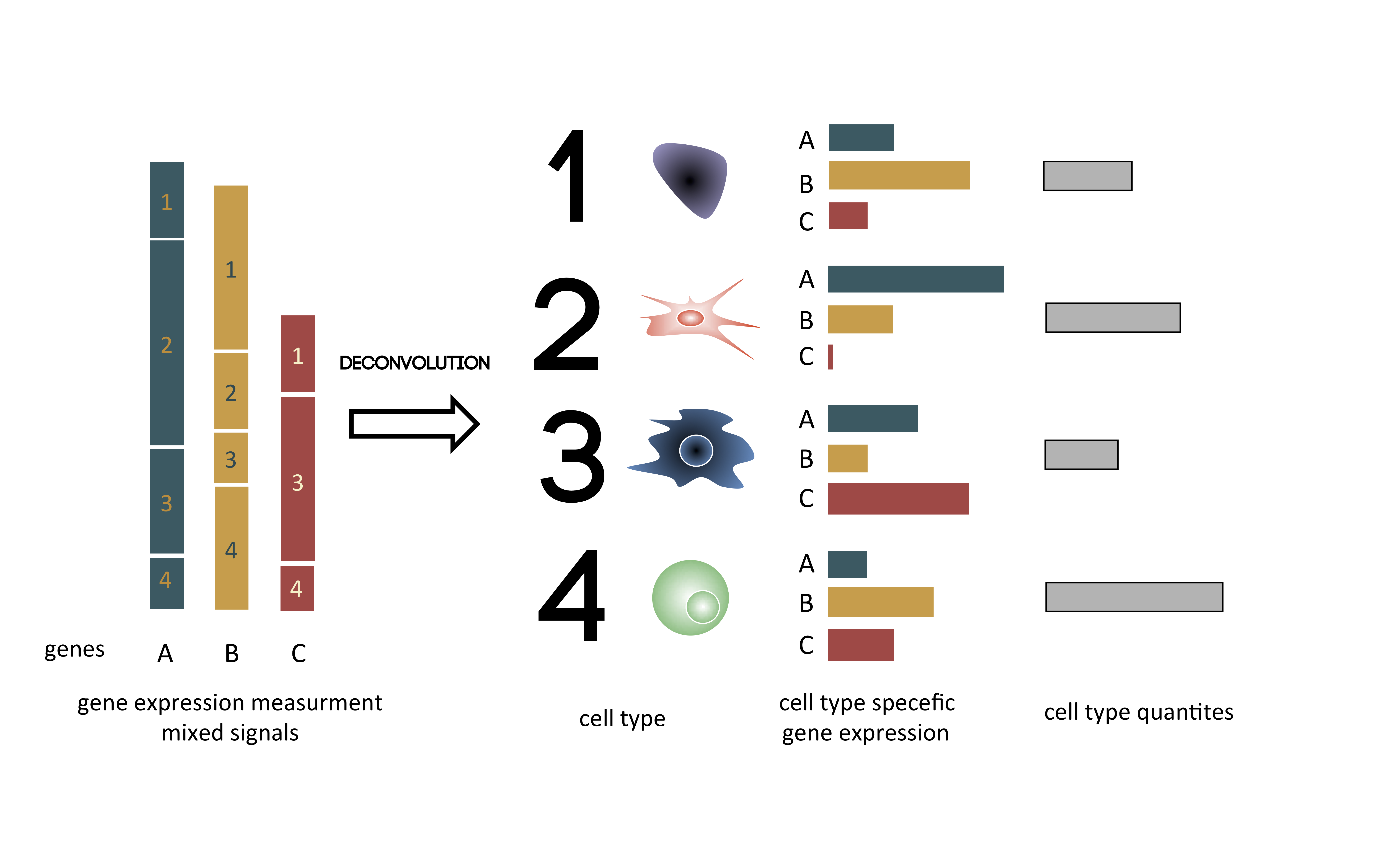
Yada is an Python library for biological cell types deconvolution. Given gene expression data, a deconvolution algorithm is capable of estimating cell type proportions in mixe of cellss. Yada is capable of deconvoluting either by using a list of marker genes or by using a complete pure gene expression matrix. Yada offers the following novelties:
- Performance: Yada’s results on benchmark datasets reached top results on a recent Dream challenge.
- Flexibility: Can be used with pure gene expression matrix or with marker-genes list only. Its core algorithm supports different sequencing platforms.
- Speed: Yada is very fast compared to other methods due to its parralel ensemble design.
- Yada is one of the few deconvolution tools that are based on Python which is the lingua franca of data scientists today.
- Availability: Can be run either as a Jupyter notebook or standalone script.
- Matrix decomposition https://en.wikipedia.org/wiki/Matrix_decomposition
- Benchmark data sets are available in the data folder (TIMER, PertU, DSA, DeconRNASeq, Abbas, BreastBlood, RatBrain, EPIC, CIBERSORT).
- Two files:
- pure.csv: pure cell genes expression file. (n genes) x (k cell types)
- mix.csv: mixtures genes expression file. (n genes) x (m mixtures).
- Gene symbols in column 1; Mixture labels in row 1.
- Tabular format with no missing entries.
- It is OK if some genes are missing from the either file.
- Data is assumed to be in non-log space (scale). If the dataset maximum expression value is less than 50, we run anti-log on all expression values.
- Yada performs a marker gene selection algorithm and therefore typically does not use all genes in the signature matrix. If this step is not needed a simple code change should comment the relevant lines.
- Browse to https://colab.research.google.com/.
- Using the Github tab select https://github.com/zurkin1/Yada.git.
- Open Yada.ipynb and follow instructions.
Table: Summary of methods for cell-type deconvolution of bulk transcriptome
Show 10202550100 entries
Search:
| name | number of sources | data | type | method | doi | author | year | proportions.in | profiles.in | application | availability | out.profiles | out.proportions | comments | category | language | citations | pop.index | published | previously.covered |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
name | number of sources | data | type | method | doi | author | year | proportions.in | profiles.in | application | availability | out.profiles | out.proportions | comments | category | language | citations | pop.index | published | previously.covered |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIBERSORT | 22 | MA | supervised | Supported vector regression | https://doi.org/10.1038/nmeth.3337 | Aaron M Newman | 2015 | FALSE | FALSE | Cancer transcriptome | http://cibersort.stanford.edu/ | FALSE | TRUE | regression | R, web tool | 343 | 85.75 | journal | TRUE | |
| ESTIMATE | 2 | MA + RNA-seq | supervised | ssGSEA | https://doi.org/10.1038/ncomms3612 | Kosuke Yoshihara | 2013 | FALSE | FALSE | Cancer transcriptome | https://sourceforge.net/projects/estimateproject/ | FALSE | TRUE | purity estimation | enrichment | R | 266 | 44.3333333333333 | journal | TRUE |
| csSAM | user-defined | MA | supervised | least-squares regression | https://doi.org/10.1038/nmeth.1439 | Shai S Shen-Orr | 2010 | TRUE | FALSE | Blood | https://github.com/shenorrLab/csSAM | TRUE | FALSE | computes DEG | regression | R | 286 | 31.7777777777778 | journal | TRUE |
| Virtual Microdissection | 14 | MA | unsupervised | multiplicative update NMF | https://doi.org/10.1038/ng.3398 | Richard A Moffitt | 2015 | TRUE | FALSE | detection of cancer and stroma in PDAC (TCGA) | NA | TRUE | TRUE | matrix factorisation | matlab | 86 | 21.5 | journal | TRUE | |
| Abbas regression | 17 | MA | supervised | linear least squares regression | https://doi.org/10.1371/journal.pone.0006098 | Alexander R. Abbas | 2009 | FALSE | TRUE | Blood | NA | FALSE | TRUE | regression | R | 207 | 20.7 | journal | TRUE | |
| MCPcounter | 10 | MA | supervised | means of marker genes | https://doi.org/10.1186/s13059-016-1070-5 | Etienne Becht | 2016 | FALSE | FALSE | Cancer transcriptome | https://github.com/ebecht/MCPcounter | FALSE | TRUE | enrichment | R | 42 | 14 | journal | TRUE | |
| PSEA | user-defined | MA | supervised | linear regression | https://doi.org/10.1038/nmeth.1710 | Alexandre Kuhn | 2011 | FALSE | TRUE | Brain tissue | https://bioconductor.org/packages/release/bioc/html/PSEA.html | FALSE | TRUE | regression | R | 96 | 12 | journal | TRUE | |
| Quadratic programming | 6 | MA | supervised | linear latent variable model solved with quadratic programming | https://doi.org/10.1371/journal.pone.0027156 | Ting Gong | 2011 | TRUE | TRUE | Blood | NA | FALSE | TRUE | regression | unknown | 76 | 9.5 | journal | TRUE | |
| Semi-supervised Nonnegative Matrix Factorization | user-defined | MA | semi-supervised | NMF minimizing the Kullback-Leibler divergence on pre-selected genes and with pure proportions | https://doi.org/10.1016/j.meegid.2011.08.014 | Renaud Gaujoux | 2012 | TRUE | TRUE | Blood | https://web.cbio.uct.ac.za/~renaud/CRAN/web/CellMix/ | TRUE | TRUE | matrix factorisation | R | 61 | 8.71428571428571 | journal | TRUE | |
| ssGSEA applied to renal cell carcinoma | 30 | RNA-seq | supervised | ssGSEA | https://doi.org/10.1186/s13059-016-1092-z | Yasin Şenbabaoğlu | 2016 | FALSE | FALSE | Cancer transcriptome | NA | FALSE | TRUE | used Bindea et al. Signatures, validated with FACS, CNV and methymome | enrichment | R | 40 | 13.3333333333333 | journal | TRUE |
| DeconRNASeq | undefiend | RNA-seq | supervised | linear latent variable model solved with quadratic programming | https://doi.org/10.1093/bioinformatics/btt090 | Ting Gong | 2013 | TRUE | TRUE | Tissue mixtures | https://www.bioconductor.org/packages/release/bioc/html/DeconRNASeq.html | FALSE | TRUE | regression | R | 52 | 8.66666666666667 | journal | TRUE | |
| DSA | user-defined | MA | supervised | linear model solved with quadratic programming. | https://dx.doi.org/10.1186%2F1471-2105-14-89 | Yi Zhong | 2013 | FALSE | TRUE* | Cancer transcriptome | https://github.com/zhandong/DSA | TRUE | TRUE | regression | R | 52 | 8.66666666666667 | journal | TRUE | |
| DECONVOLUTE | user-defined | MA | supervised | system of linear equations | https://doi.org/10.1073/pnas.1832361100 | Peng Lu | 2003 | TRUE | TRUE | yeast cell cycle | broken link | FALSE | TRUE | regression | Java 2 | 135 | 8.4375 | journal | TRUE | |
| ISOpure | 2 | MA | supervised | maximum a posteriori(MAP) estimation of multinomial distribution | https://doi.org/10.1186/gm433 | Gerald Quon | 2013 | FALSE | TRUE | Cancer transcriptome | https://qlab.faculty.ucdavis.edu/isopure/ | TRUE | TRUE | purity estimation | probabilistic | matlab, R | 44 | 7.33333333333333 | journal | TRUE |
| xCell | 64 | MA + RNA-seq | supervised | ssGSEA+ spillover compensation | https://dx.doi.org/10.1186%2Fs13059-017-1349-1 | Dvir Aran | 2017 | FALSE | FALSE | Cancer transcriptome | http://xcell.ucsf.edu/; https://github.com/dviraran/xCell | FALSE | TRUE | deep deconvolution | enrichment | R, web tool | 15 | 7.5 | journal | FALSE |
| CellCODE | user-defined | MA | semi-supervised | robust latent variable | https://doi.org/10.1093/bioinformatics/btv015 | Maria Chikina | 2015 | TRUE | TRUE* | Blood | http://www.pitt.edu/~mchikina/CellCODE/ | TRUE | TRUE | goal improve DEG, plugs marker-genes based covariated to the SVD | matrix factorisation | R- C-C++- Fortran | 28 | 7 | journal | TRUE |
| DCQ | undefiend | RNA-seq | supervised | regularized regression model | https://dx.doi.org/10.1002%2Fmsb.134947 | Zeev Altboum | 2014 | TRUE | TRUE | Mice blood under flu infection | http://www.dcq.tau.ac.il/ | FALSE | TRUE | comparing conditions (normal vs infection) | regression | web tool | 32 | 6.4 | journal | TRUE |
| DeMix | 2 | MA | supervised | Maximul likelihood estimate | https://doi.org/10.1093/bioinformatics/btt301 | Jaeil Ahn | 2013 | FALSE | TRUE | Cancer purity | http://odin.mdacc.tmc.edu/∼wwang7/DeMix.html. | TRUE | TRUE | on log transformed data | probabilistic | C, R | 38 | 6.33333333333333 | journal | TRUE |
| Statistical expression deconvolution | 2 | MA | supervised | linear equations | https://doi.org/10.1093/bioinformatics/btq097 | Jennifer Clarke | 2010 | FALSE | TRUE | Cancer xenografts | NA | FALSE | TRUE | regression | unknown | 53 | 5.88888888888889 | journal | TRUE | |
| DSection | user-defined | MA | supervised | Bayesian MCMC-based model | https://doi.org/10.1093/bioinformatics/btq406 | Timo Erkkilä | 2010 | TRUE | FALSE | Tissue mixtures | http://informatics.systemsbiology.net/DSection | TRUE | FALSE | probabilistic | matlab | 52 | 5.77777777777778 | journal | TRUE | |
| Nanodissection | undefined | MA | supervised | iterative SVM | https://doi.org/10.1101/gr.155697.113 | Wenjun Ju | 2013 | FALSE | TRUE | Chronic kidney disease (Cell lineages) | http://nano.princeton.edu/ | FALSE | TRUE | identifies genes | regression | web tool | 33 | 5.5 | journal | TRUE |
| TIMER | 6 | MA + RNA-seq | supervised | constrained linear regression | https://doi.org/10.1186/s13059-016-1028-7 | Bo Li | 2013 | FALSE | TRUE | Cancer transcriptome | http://cistrome.org/TIMER/ | FALSE | TRUE | made for TCGA, other application need adaptation | regression | web tool | 33 | 5.5 | journal | TRUE |
| Direct method | SM | MA | unsupervised | non negative least squares and decorellation | https://www.ncbi.nlm.nih.gov/pubmed/11473019 | David Venet | 2001 | TRUE | FALSE | cander and normal tissue | NA | TRUE | TRUE | matrix factorisation | unknown | 96 | 5.33333333333333 | journal | TRUE | |
| SPEC | user-defined | MA | supervised | enrichment score | https://doi.org/10.1186/1471-2105-12-258 | Christopher R Bolen | 2011 | FALSE | TRUE* | Blood | http://clip.med.yale.edu/SPEC/ | FALSE | TRUE | enrichment | R | 39 | 4.875 | journal | TRUE | |
| PERT | 11 | MA | supervised | Non-negative maximum likelihood model with adjustement for perturbations | https://doi.org/10.1371/journal.pcbi.1002838 | Wenlian Qiao | 2012 | TRUE | TRUE | blood | https://github.com/gquon/PERT | TRUE | TRUE | probabilistic | octave | 33 | 4.71428571428571 | journal | TRUE | |
| deconf | user-defined | MA | unsupervised | Least squares non-negative matrix factorization algorithm | https://doi.org/10.1186/1471-2105-11-27 | Dirk Repsilber | 2010 | FALSE | FALSE | Blood | https://static-content.springer.com/esm/art%3A10.1186%2F1471-2105-11-27/MediaObjects/12859_2009_3484_MOESM1_ESM.ZIP | TRUE | TRUE | matrix factorisation | R | 41 | 4.55555555555556 | journal | TRUE | |
| CTen | undefined | MA | supervised | GSEA | https://doi.org/10.1186/1471-2164-13-460 | Jason E Shoemaker | 2012 | FALSE | FALSE | Infected lung tissue | http://www.influenza-x.org/~jshoemaker/cten/ | TRUE | FALSE | first enrichment based | enrichment | web tool | 31 | 4.42857142857143 | journal | TRUE |
| Mixture models | 2 | MA | supervised | methods of moments procedures and the expectation–maximization algorithm | https://doi.org/10.1093/bioinformatics/bth139 | Debashis Ghosh | 2004 | FALSE | TRUE | Cancer transcriptome | broken link | TRUE | TRUE | improve DEG | probabilistic | R | 66 | 4.4 | journal | TRUE |
| CAM | 10 | MA | unsupervised | convex analysis of mixtures | https://doi.org/10.1038/srep18909 | Niya Wang | 2016 | FALSE | FALSE | yeast cell cycle | http://mloss.org/software/view/437, | TRUE | TRUE | partial profiles | convex hull | R-java | 12 | 4 | journal | TRUE |
| ISOLATE | undefined | MA | supervised | Latent Dirichlet Allocation (LDA) | https://dx.doi.org/10.1093%2Fbioinformatics%2Fbtp378 | Gerald Quon | 2009 | FALSE | FALSE | Cancer transcriptome | https://qlab.faculty.ucdavis.edu/isolate/ | TRUE | TRUE | designed for detection of site of origin | matrix factorisation | matlab | 37 | 3.7 | journal | TRUE |
| UNDO | 2 | MA | unsupervised | matrix inversion | https://doi.org/10.1093/bioinformatics/btu607 | Niya Wang | 2014 | FALSE | FALSE | Cancer transcriptome | https://www.bioconductor.org/packages/release/bioc/html/UNDO.html | TRUE | TRUE | matrix factorisation | R | 18 | 3.6 | journal | TRUE | |
| In silico microdissection | 3 | MA | unsupervised | Gaussian Mixture Model | https://doi.org/10.1186/1471-2105-6-54 | Harri Lähdesmäki | 2005 | FALSE | FALSE | In vitro tissue mixtures | NA | TRUE | TRUE | probabilistic | unknown | 45 | 3.21428571428571 | journal | TRUE | |
| BioQC | 150 | MA + RNA-seq | supervised | Wilcoxon-Mann-Whitney test | https://doi.org/10.1186/s12864-017-3661-2 | Jitao David Zhang | 2017 | FALSE | FALSE | Gene expression | https://www.bioconductor.org/packages/release/bioc/html/BioQC.html | FALSE | FALSE | heterogeneity detection | enrichment | R | 6 | 3 | journal | TRUE |
| Self-directed Method for Cell-Type Identification | undefined | MA | unsupervised | non-negative least squares, Kullback-Leibler divergence | https://doi.org/10.1371/journal.pcbi.1003189 | Neta S. Zuckerman | 2013 | FALSE | TRUE | Cancer transcriptome | NA | TRUE | TRUE | matrix factorisation | matlab | 18 | 3 | journal | TRUE | |
| Computational expression deconvolution | undefined | MA | supervised | linear equations with simulated annealing | https://doi.org/10.1186/1471-2105-7-328 | Min Wang | 2006 | TRUE | FALSE | Murine mammary gland | NA | FALSE | TRUE | regression | unknown | 36 | 2.76923076923077 | journal | TRUE | |
| Electronical substraction | 2 | MA | supervised | system of linear equations | https://doi.org/10.1093/bioinformatics/btm508 | Mark M. Gosink | 2007 | FALSE | TRUE* | Infected macrophages | NA | TRUE | TRUE | pays attention to rare cell types | regression | unknown | 30 | 2.5 | journal | TRUE |
| EPIC | 8 | RNA-seq | supervised | weighted constrained least square optimization | https://dx.doi.org/10.7554%2FeLife.26476 | Julien Racle | 2017 | FALSE | FALSE | Cancer transcriptome | https://github.com/GfellerLab/EPIC | FALSE | TRUE | absolute quantities, requires TPM normalized data | regression | R | 4 | 2 | journal | FALSE |
| MMAD | unknown | MA | BOTH | constrained regression with corrected AIC parameter fit | https://doi.org/10.1093/bioinformatics/btt566 | David A. Liebner | 2013 | FALSE | TRUE | in vitro tissue mlxtures | http://sourceforge.net/projects/mmad/ | TRUE | TRUE | regression | matlab | 11 | 1.83333333333333 | journal | TRUE | |
| Immune Quant | user-defined | undefined | supervised | DCQ algorithm adapted to human | https://doi.org/10.1093/bioinformatics/btw535 | Amit Frishberg | 2016 | TRUE | TRUE | Human tissues | http://csgi.tau.ac.il/ImmQuant/ | FALSE | TRUE | regression | web tool | 5 | 1.66666666666667 | journal | TRUE | |
| VoCAL | user-defined | MA, GWAS | supervised | linear regression | https://doi.org/10.1371/journal.pcbi.1004856 | Yael Steuerman | 2016 | TRUE | TRUE | Lung tissue | https://cran.r-project.org/web/packages/ComICS/index.html | FALSE | TRUE | retruns association with eQTLS | regression | R | 5 | 1.66666666666667 | journal | TRUE |
| CellMapper | user-defined | MA | semi-supervised | SVD | https://doi.org/10.1186/s13059-016-1062-5 | Bradlee D. Nelms | 2016 | FALSE | TRUE* | Brain tissue | http://bioconductor.org/packages/release/bioc/html/CellMapper.html | TRUE | FALSE | ranks genes, computes p-value of specificity | matrix factorisation | R | 5 | 1.66666666666667 | journal | TRUE |
| Estimation of immune cell content | 8 | scRNA-seq | supervised | CIBERSORT using scRNA-seq basis matrix | https://doi.org/10.1038/s41467-017-02289-3 | Max Schelker | 2017 | FALSE | FALSE | Cancer transcriptome | NA | FALSE | TRUE | regression | unknown | 3 | 1.5 | journal | FALSE | |
| contamDE | 2+ | RNA-seq | supervised | empirical Bayes estimate of the negative binomial dispersion | https://doi.org/10.1093/bioinformatics/btv657 | Qi Shen | 2016 | TRUE | TRUE | Tumor purity | https://github.com/zhanghfd/contamDE/ | TRUE | TRUE | computes differential gene expression staistics | probabilistic | R | 4 | 1.33333333333333 | journal | TRUE |
| GSVA scores | 6 | RNA-seq | supervised | GSVA | https://doi.org/10.1158/1078-0432.CCR-17-3509 | David Tamborero | 2018 | FALSE | FALSE | Cancer transcriptome | NA | FALSE | TRUE | enrichment | unknown | 1 | 1 | journal | FALSE | |
| Enumerateblood | 6 | MA | supervised | multi-response Gaussian model trained | https://doi.org/10.1186/s12864-016-3460-1 | Casey P. Shannon | 2017 | FALSE | FALSE | Blood gene expression | https://github.com/cashoes/enumerateblood | TRUE | TRUE | speciffic to Affymetrix Gene ST, pre-trained | probabilistic | R | 2 | 1 | journal | TRUE |
| CoD | 207 | RNA-seq | supervised | DCQ algorithm + random forest classifier | https://doi.org/10.1093/bioinformatics/btv498 | Amit Frishberg | 2015 | TRUE | FLASE | Mice diseased tissues | http://www.csgi.tau.ac.il/CoD/ | FALSE | TRUE | distinguish cell types important in a disease | regression | web tool | 4 | 1 | journal | TRUE |
| ImmunoStates | 20 | MA | supervised | regression (previously published) | https://doi.org/10.1101/206466 | Francesco Vallania | 2017 | FALSE | FALSE | Blood, solid tissue, disease | NA | FALSE | TRUE | regression | R | 1 | 0.5 | bioRxiv | FALSE | |
| quanTIseq | 11 | RNA-seq + Images | supervised | constrained least squares regression | https://doi.org/10.1101/223180 | Francesca Finotello | 2017 | FALSE | FALSE | Cancer transcriptome | http://icbi.at/software/quantiseq/doc/index.html | FALSE | TRUE | absolute quantities from images, strating with raw data, returns cell densities | regression | web tool | 1 | 0.5 | bioRxiv | FALSE |
| SMC | user-defined | MA | unsupervised | Bayesian inference with sequential monte carlo samplers | https://doi.org/10.1371/journal.pone.0186167 | Oyetunji E. Ogundijo | 2017 | FALSE | FLASE | Tissue mixtures | https://github.com/moyanre/smcgenedeconv | TRUE | TRUE | DEG analysis | probabilistic | matlab | 1 | 0.5 | journal | FALSE |
| Modular discrimination index | 5 | MA + RNA-seq | supervised | correlation-based score | https://doi.org/10.1371/journal.pone.0169271 | Gabriele Pollara | 2017 | FALSE | FALSE | Skin tuberculosis | https://github.com/MJMurray1/MDIScoring | FALSE | TRUE | optimisation of signature genes | enrichment | R | 1 | 0.5 | journal | FALSE |
| Robust Computational Reconstitution | SM | MA | supervised | trimmed least modulus (L1) regression. | https://doi.org/10.1186/1471-2105-7-369 | Martin Hoffmann | 2006 | TRUE | TRUE | Synovial tissue (cell types in silico) | NA | FALSE | TRUE | regression | unknown | 6 | 0.461538461538462 | journal | TRUE | |
| Statical mechanics approach | 2 | undefined | unsupervised | Bayesian model with MCMC sampling assuming Gaussian distribution | https://arxiv.org/abs/1210.7508v1 | Nico Riedel | 2013 | FALSE | FALSE | Udefined | NA | TRUE | TRUE | theoretical solution | probabilistic | unknown | 2 | 0.333333333333333 | arXiv | FALSE |
| MHMM | user-defined | MA | unsupervised | Hidden state markov model | https://doi.org/10.1089/cmb.2006.13.1749 | Sushmita Roy | 2006 | TRUE | FLASE | Yeast cell cycle | NA | TRUE | TRUE | deaing with missign values and time dependency | probabilistic | unknown | 4 | 0.307692307692308 | journal | TRUE |
| MySort | 22 | MA | supervised | v -Support Vector Regression | https://doi.org/10.1186/s12859-018-2069-6 | Shu-Hwa Chen | 2018 | FALSE | FALSE | Blood | https://testtoolshed.g2.bx.psu.edu/repository?repository_id=6e9a9ab163e578e0&changeset_revision=e3afe097e80a | FALSE | TRUE | Galaxy platform pluggable tool | regression | R, web tool | 0 | 0 | journal | FALSE |
| ADVOCATE | 2 | RNA-seq | supervised | trained Gaussian-mixture mode | https://doi.org/10.1101/288779 | Jing He | 2018 | FALSE | TRUE | Cancer transcriptome | NA | TRUE | TRUE | probabilistic | R | 0 | 0 | bioRxiv | FALSE | |
| DTD | user-defined | scRNA-seq | supervised | optimising loss-function of penalized least-squares regression | https://arxiv.org/abs/1801.08447v1 | Franziska Görtler | 2018 | TRUE | TRUE | Cancer transcriptome | NA | FALSE | TRUE | regression | unknown | 0 | 0 | bioRxiv | FALSE | |
| CellDistinguisher | user-defined | MA + RNA-seq | unsupervised | topic modelling + onvex hull / NMF | https://doi.org/10.1371/journal.pone.0193067 | Lee A. Newberg | 2018 | FALSE | FALSE | yeast cell cycle | https:// github.com/GeneralElectric/CcellDdistinguisher | TRUE | TRUE | partial profiles | convex hull | R | 0 | 0 | journal | FALSE |
| dtangle | user-defined | MA + RNA-seq | supervised | linear regression | https://doi.org/10.1101/290262 | Gregory J. Hunt | 2018 | FALSE | TRUE | Blood | https://cran.r-project.org/package=dtangle | FALSE | TRUE | regression | R | 0 | 0 | bioRxiv | FALSE | |
| DeconICA | 100 | MA + RNA-seq | unsupervised | ICA + post identification: enrichment | https://doi.org/10.5281/zenodo.1250069 | Urszula Czerwinska | 2018 | FALSE | FALSE | Cancer transcriptome | https://urszulaczerwinska.github.io/DeconICA/ | TRUE | TRUE | metagene profiles | matrix factorisation | R, matlab | 0 | 0 | NA | FALSE |
| DemixT | 3 | MA + RNA-seq | supervised | Maximal likelihood estimate | https://doi.org/10.1101/146795 | Zeya Wang | 2017 | FALSE | TRUE | Cancer transcriptome | https://github.com/wwylab/DeMixT | TRUE | TRUE | probabilistic | R | 0 | 0 | bioRxiv | FALSE | |
| Post‐modified non‐negative matrix factorization | user-defined | RNA-seq | unsupervised | NMF using alternating least square | https://doi.org/10.1002/cem.2929 | Yuan Liu | 2017 | FALSE | FALSE | Cancer transcriptome | NA | TRUE | TRUE | DEG analysis | matrix factorisation | matlab | 0 | 0 | journal | FALSE |
| Infino | user-defined | RNA-seq | supervised | Bayesian inference with a generative model | https://doi.org/10.1101/221671 | Maxim E Zaslavsky | 2017 | FALSE | TRUE | Cancer transcriptome | https://github.com/hammerlab/infino | TRUE | TRUE | specalised in deep deconvolution | probabilistic | Stan | 0 | 0 | bioRxiv | FALSE |
| ImSig | 11 | RNA-seq | supervised | correlation-based score | https://doi.org/10.1101/077487 | Ajit Johnson Nirmal | 2016 | FALSE | FALSE | Cancer transcriptome | NA | FALSE | TRUE | enrichment | unknown | 0 | 0 | bioRxiv | FALSE | |
| TEMT | 2 | RNAseq | supervised | generative mixture model | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-14-S5-S11 | Yi Li | 2013 | TRUE | TRUE | in vitro cell mixtures | https://github.com/uci-cbcl/TEMT | FALSE | TRUE | use reads | probabilistic | python | 0 | 0 | journal | TRUE |
Showing 1 to 64 of 64 entries
The most popular language of implementation of published methods is R (49.2 %), followed by Matlab (11.11%), only one tool so far was published in Python.