A Multiscale Approach for Whole-Slide Image Segmentation of five Tissue Classes in Urothelial Carcinoma Slides
This is the source code described in the paper "A Multiscale Approach for Whole-Slide Image Segmentation of five Tissue Classes in Urothelial Carcinoma Slides".
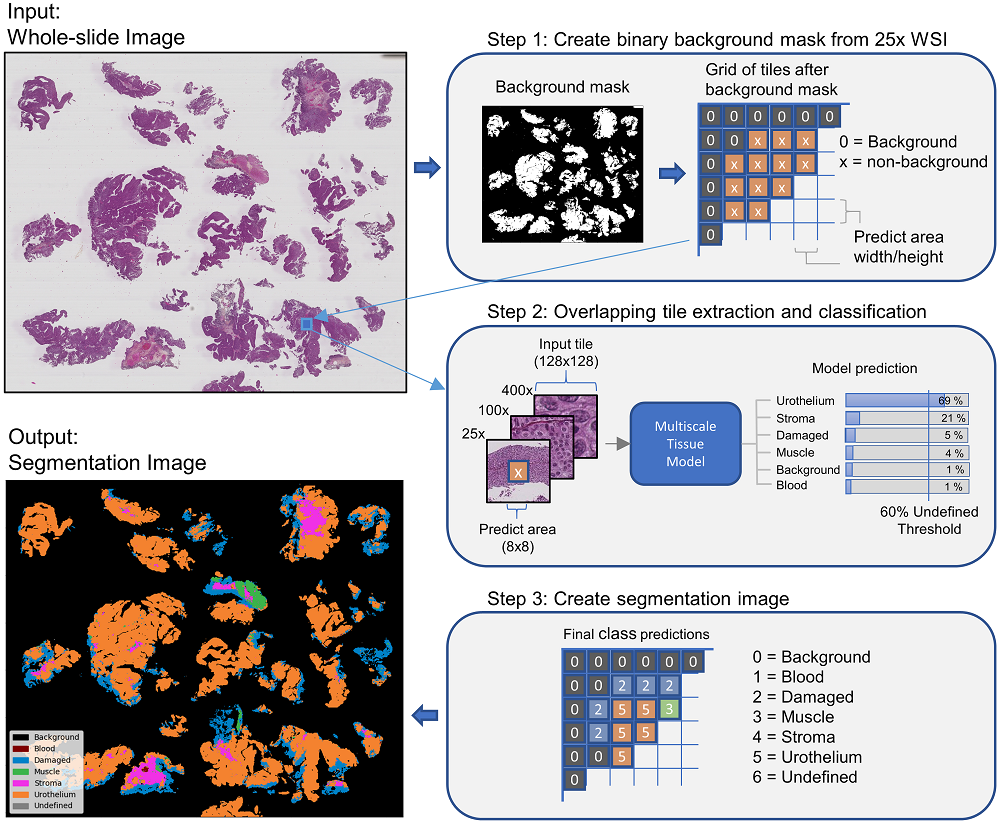
In pathology labs worldwide, we see an increasing number of tissue samples that need to be assessed without the same increase in the number of pathologists. Computational pathology, where digital scans of histological samples called whole-slide images (WSI) are processed by computational tools, can be of help for the pathologists and is gaining research interests. Most research effort has been given to classify slides as being cancerous or not, localization of cancerous regions, and to the “big-four” in cancer: breast, lung, prostate, and bowel. Urothelial carcinoma, the most common form of bladder cancer, is expensive to follow up due to a high risk of recurrence, and grading systems have a high degree of inter- and intra-observer variability. The tissue samples of urothelial carcinoma contain a mixture of damaged tissue, blood, stroma, muscle, and urothelium, where it is mainly muscle and urothelium that is diagnostically relevant. A coarse segmentation of these tissue types would be useful to i) guide pathologists to the diagnostic relevant areas of the WSI, and ii) use as input in a computer-aided diagnostic (CAD) system. However, little work has been done on segmenting tissue types in WSIs, and on computational pathology for urothelial carcinoma in particular. In this work, we are using convolutional neural networks (CNN) for multiscale tile-wise classification and coarse segmentation, including both context and detail, by using three magnification levels: 25x, 100x, and 400x. 28 models were trained on weakly labeled data from 32 WSIs, where the best model got an F1-score of 96.5% across six classes. The multiscale models were consistently better than the single-scale models, demonstrating the benefit of combining multiple scales. No tissue-class ground-truth for complete WSIs exist, but the best models were used to segment seven unseen WSIs where the results were manually inspected by a pathologist and are considered as very promising.
The Python program in the Preprocessing folder is used to find all tiles within a region of interest (ROI) and save the coordinates in pickle files. These pickle files is then used to train the models in the 'tissue-segmentation-model' folder. The code is created to read XML files generated using Aperio ImageScope. Aperio ImageScope has a drawing tool which can be used to annotate ROIs in the WSI. Only one tissue type can be annotated in a WSI at a time. To annotate multiple tissue types in the same WSI, copy the WSI to create one version for each tissue type.
In the root directory of the preprocessing code, create a folder 'input/'. In this foler, create one folder for each tissue type to extract, e.g., 'input/Urothelium/' and 'input/Stroma/'. For each tissue folder, create a folder with the WSI name and place your WSI and XML file inside.
Requirements:
python==3.6.7
pyvips==2.1.12
In the root directory, create the folders 'Coordinate_dicts_files/tissue_groundtruth_training/' and 'Coordinate_dicts_files/tissue_groundtruth_test/'. Take the resulting pickle-files from the preprocessing program and place them into the training and test folders.
The main file contain five different 'modes' to choose from. To enable a mode, set the top variale MODE_* to True. To run the main file, add the argument "True" or "False". "True" means you are starting a new model, and a new folder will be created. "False" means you are not starting a new model, and want to resume an existing model. If you have multiple models, use the variable CONTINUE_FROM_MODEL to specify which model to start from. By default, CONTINUE_FROM_MODEL is set to "last", which will resume the most recent model.
The five modes are:
- Train a tissue model using cross-validation
- Train a tissue model with only a training set (this can be used after cross-validation for retraining a model on all available data)
- Train a tissue model using training and validation dataset
- Test a tissue model on the test dataset
- Use a model to predict tissue on new WSIs (this mode is used to generate the tissue-map on the above image)
The code contain a large list of options. From the main file it is possible to choose which base model to use, freeze/unfreeze base model, dropout rate, number of neurons in the classifier layers, how many scales to use and which scales to use. There is a debug mode where only a small dataset is used. For data augmentation, it is possible to choose which tissue classes to augment, and how many times to augment the class. If the training is stopped due to e.g. a power outage, it is possible to continue training from the last epoch, this also workes if training multiple models or cross-validation. It is possible to enter a list of hyperparameters, e.g. learning rate = [0.1, 0.01] dropout_rate = [0.1, 0.4], and the program will train all possible combinations of the hyperparameters. The program have the option to save learning curves, confusion matrices, classification reports, misclassified images and logs.
Requirements:
python==3.6.7
numpy==1.18.5
scikit-image==0.17.2
pyvips==2.1.12
scipy==1.4.1
tensorflow==1.13.1
keras==2.2.4
matplotlib==3.1.0
PIL==6.0.0
https://journals.sagepub.com/doi/full/10.1177/1533033820946787
The code is released free of charge as open-source software under the GPL-3.0 License. Please cite our paper when you have used it in your study.
Rune Wetteland, Kjersti Engan, Trygve Eftestøl, Vebjørn Kvikstad, and Emiel AM Janssen.
@article{wetteland2020multiscale,
title={A Multiscale Approach for Whole-Slide Image Segmentation of five Tissue Classes in Urothelial Carcinoma Slides},
author={Wetteland, Rune and Engan, Kjersti and Eftest{\o}l, Trygve and Kvikstad, Vebj{\o}rn and Janssen, Emiel AM},
journal={Technology in Cancer Research \& Treatment},
volume={19},
pages={1533033820946787},
year={2020},
publisher={SAGE Publications Sage CA: Los Angeles, CA}
}