The goal of spatialsample is to provide functions and classes for spatial resampling to use with rsample, including:
The implementation of more spatial resampling approaches is planned.
Like rsample, spatialsample provides building blocks for creating and analyzing resamples of a spatial data set but does not include code for modeling or computing statistics. The resampled data sets created by spatialsample are efficient and do not have much memory overhead.
You can install the released version of spatialsample from CRAN with:
install.packages("spatialsample")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("tidymodels/spatialsample")The most straightforward spatial resampling strategy with the lightest
dependencies is spatial_clustering_cv(), which uses k-means clustering
to identify cross-validation folds:
library(spatialsample)
data("ames", package = "modeldata")
set.seed(1234)
folds <- spatial_clustering_cv(ames, coords = c("Latitude", "Longitude"), v = 5)
folds
#> # 5-fold spatial cross-validation
#> # A tibble: 5 x 2
#> splits id
#> <list> <chr>
#> 1 <split [2332/598]> Fold1
#> 2 <split [2187/743]> Fold2
#> 3 <split [2570/360]> Fold3
#> 4 <split [2118/812]> Fold4
#> 5 <split [2513/417]> Fold5In this example, the ames data on houses in Ames, IA is resampled with
v = 5; notice that the resulting partitions do not contain an equal
number of observations.
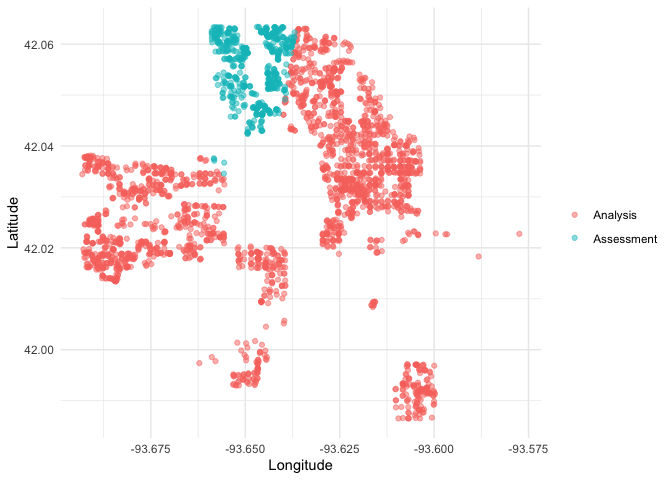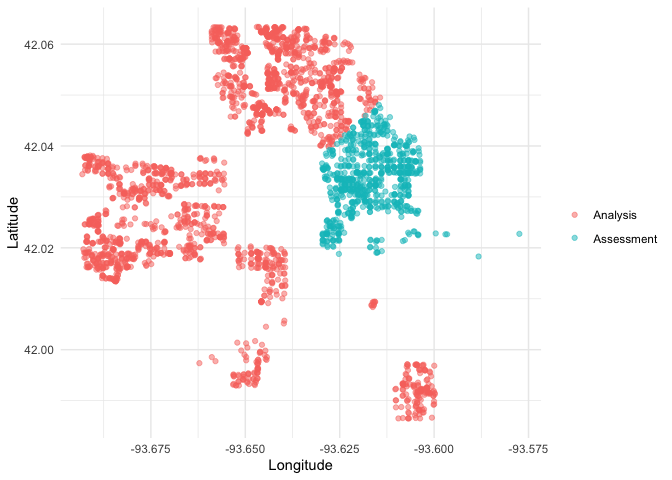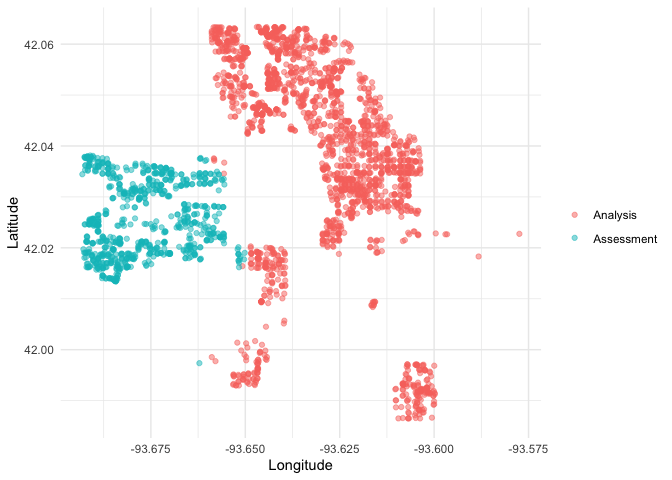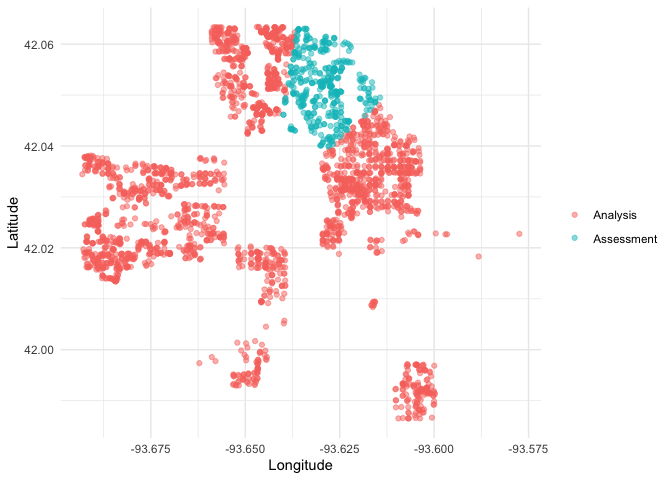
We can create a helper plotting function to visualize the five folds.
library(ggplot2)
library(purrr)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
plot_splits <- function(split) {
p <- analysis(split) %>%
mutate(analysis = "Analysis") %>%
bind_rows(assessment(split) %>%
mutate(analysis = "Assessment")) %>%
ggplot(aes(Longitude, Latitude, color = analysis)) +
geom_point(alpha = 0.5) +
labs(color = NULL)
print(p)
}
walk(folds$splits, plot_splits)This project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
-
For questions and discussions about tidymodels packages, modeling, and machine learning, please post on RStudio Community.
-
If you think you have encountered a bug, please submit an issue.
-
Either way, learn how to create and share a reprex (a minimal, reproducible example), to clearly communicate about your code.
-
Check out further details on contributing guidelines for tidymodels packages and how to get help.