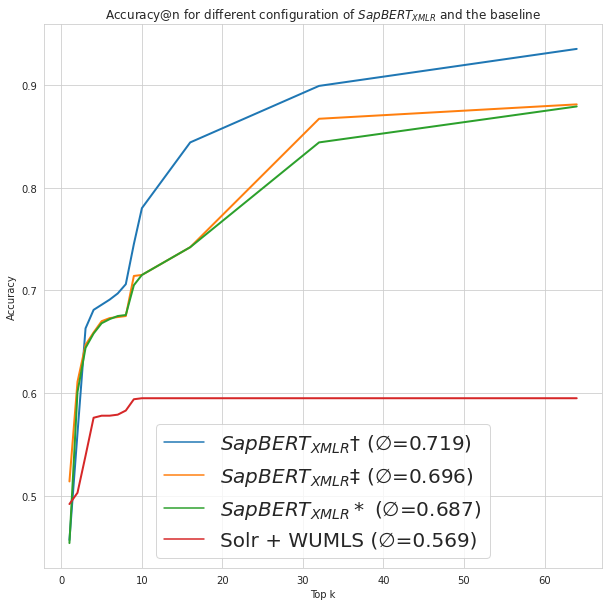
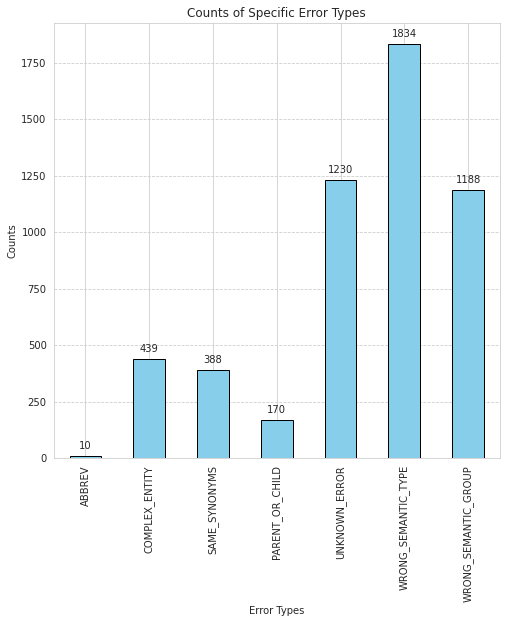
In the field of biomedical natural language processing, medical concept normalization is a crucial task for accurately mapping mentions of concepts to a large knowledge base. However, this task becomes even more challenging in low-resource settings, where limited data and resources are available. In this thesis, I explore the challenges of medical concept normalization in a low-resource setting. Specifically, I investigate the shortcomings of current medical concept normalization methods applied to German lay texts. Since there is no suitable dataset available, a dataset consisting of posts from a German medical online forum is annotated with concepts from the Unified Medical Language System. The experiments demonstrate that multilingual Transformer-based models are able to outperform string similarity methods. The use of contextual information to improve the normalization of lay mentions is also examined, but led to inferior results. Based on the results of the best performing model, I present a systematic error analysis and lay out potential improvements to mitigate frequent errors.
The code for the main experiment can be found in sapbert_xlmr_tlc_create_embeddings.ipynb and sapbert_xlmr_tlc_ger_similarity_search.ipynb. The multilingual SapBERT model is used to embed mentions from TLC-UMLS and concepts from UMLS. The closest concept for each mention is found using cosine similarity.
The experiment to train a Sentence Cross-Encoder to re-rank candidate concepts can be found in train_cross_encoder_xmen.ipynb