- EXAMM and EXA-GP Overview
- Installation and Setup
- Quickstart
- Managing Datasets
- Running EXAMM and EXA-GP
- Tracking and Managing Evolved Networks
- Using Evolved Neural Networks for Inference
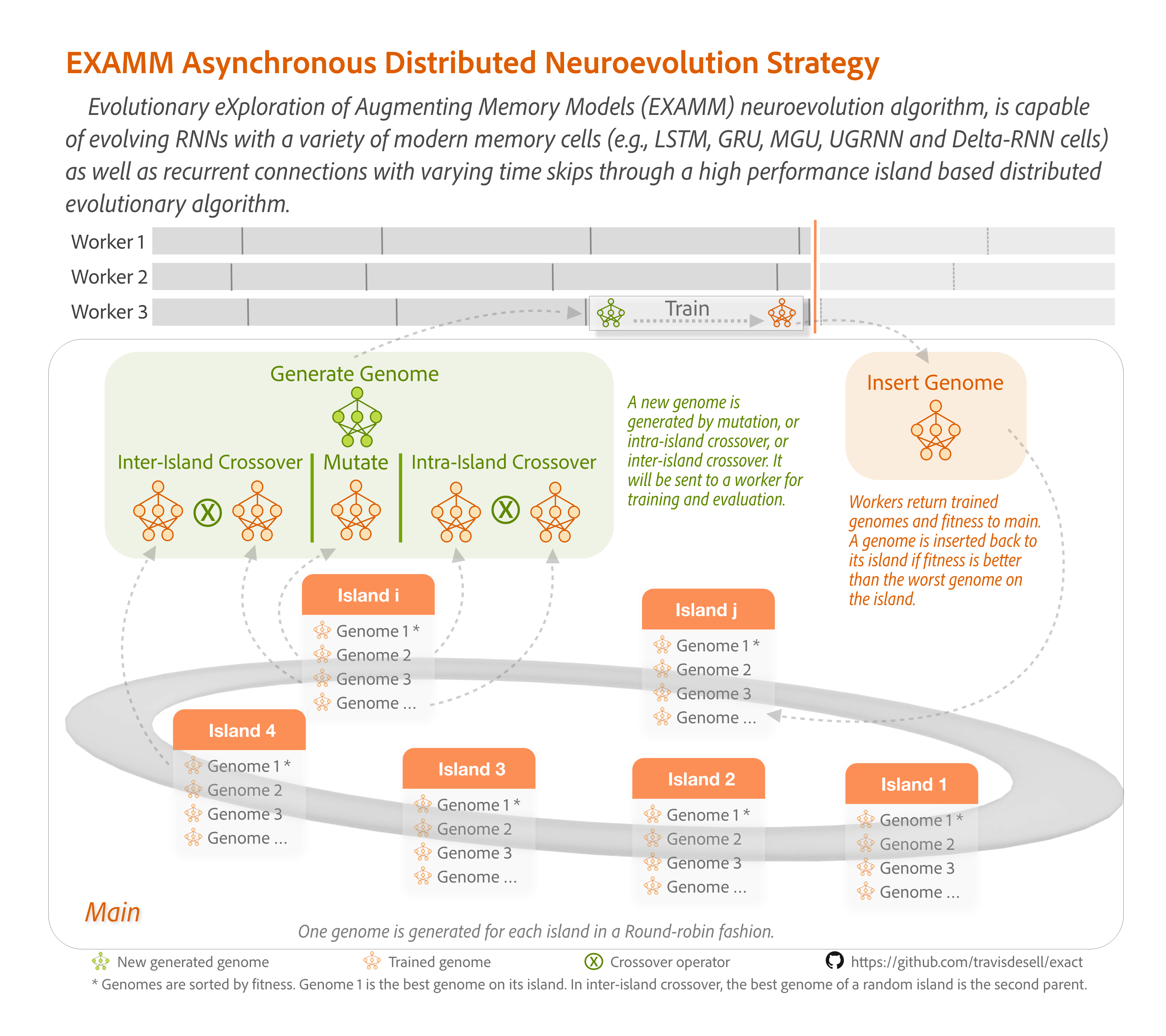
EXAMM (Evolutionary eXploration of Augmenting Memory Models) is a neuroevolution (evolutionary neural architecture search) algorithm which automates the design and training of recurrent neural networks (RNNs) for time series forecasting. EXAMM uses a constructive evolutionary process which evolves progressively larger RNNs by a set of mutation and crossover operations. EXAMM is a fine-grained neuroevolution algorith, operating at the level of individual nodes and edges which allows for extremely efficient and minimal networks. It utilizes a library of various modern memory cells (LSTM, GRU, MGU, UGRNN, and Delta-RNN) 1 and can establish recurrent connections with varying time skips for improved learning and forecasting 2. It also uses a Lamarckian weight inheritance strategy, allowing generated networks to re-use weights of their parents to reduce the amount of training by backpropagation required 3.
EXAMM has since been extended to the Evolutionary Exploration of Augmenting Genetic Programs (EXA-GP) algorithm, which replaces the memory cells of EXAMM with basic genetic programming (GP) operations (e.g., sum, product, sin, cos, tanh, sigmoid, inverse). EXA-GP has been shown to generate compact genetic programs (multivariate functions) for time series forecasting which can outperform the RNNs evolved by EXAMM while at the same time being more interpretable45.
Implemented in C++, EXAMM and EXA-GP are designed for efficient CPU-based computation (which for time series forecasting RNNs are typically more performant than GPUs) and offers excellent scalability due to its asynchronous island based distributed strategy (see above) with repopulation events which prune evolutionary dead ends to improve perforance6. They employ a distributed architecture where worker processes handle RNN training while a main process manages population evolution and orchestrates the overall evolutionary process. This allows for better performance via either multithreaded execution or distributed execution on high performance computing clusters via the message passing interface (MPI).
EXAMM and EXA-GP have been designed to have a fairly minimal set of requirements, and we recommend using either OSX or Linux. For Windows users, we recommend using Windows Subsystem for Linux (WSL) to run EXAMM or EXA-GP in a linux VM. EXAMM/EXA-GP use CMake to create a makefile for building (this can potentially also be used to make a visual studio project, however we have not tested this).
For OSX we recommend using Homebrew to handle installing packages, for Linux please use your package manager of choice. Installing all required libraries below (or their linux versions) should be sufficient to compile EXAMM/EXA-GP:
xcode-select --install
brew install cmake
brew install mysql
brew install open-mpi
brew install libtiff
brew install libpng
brew install clang-formatThe following is for internal use on RIT's high performance computing cluster, however if your own computing cluster utilizes Spack you may find this useful.
# GCC (9.3)
spack load gcc/lhqcen5
# CMake
spack load cmake/pbddesj
# OpenMPI
spack load openmpi/xcunp5q
# libtiff
spack load libtiff/gnxev37After the above libraries have been installed and/or loaded, compiling EXAMM/EXA-GP should be as simple doing the following within your root EXAMM directory.
mkdir build
cd build
cmake ..
makeFor quick start with example datasets using basic settings, the following scripts provide examples of running EXAMM on the coal benchmark datasets provided in this repository running either the multithreaded version or the MPI version. For a deeper dive on EXAMM/EXA-GP's command line arguments please see the Running EXAMM and EXA-GP section.
# In the root directory:
sh scripts/base_run/coal_mt.sh# In the root directory:
sh scripts/base_run/coal_mpi.shEXAMM and EXA-GP are designed to use multivariate time series data as training and validation data. When EXAMM or EXA-GP generate a new recurrent neural network (RNN) or genetic program (GP), the RNN or GP is trained for a specified number of backpropagation epochs on the training data, and then the fitness of the RNN or GP is calculated by evaluating it using the validation data. Simple comma-separated value (CSV) files are used to represent th the training and validation data (examples can be found within the datasets subdirectory of the project). The first row of the CSV file should contain the column headers (without a # character), and all columns should have numerical values as data. For example:
file1.csv:
a,b,c,d
0.5,0.2,0.1,0.2
0.8,0.1,0.3,0.5
...
0.9,-0.2,0.2,0.6
file2.csv:
a,b,c,d
0.7,-0.2,0.7,0.3
0.6,-0.1,0.5,0.4
...
0.4,0.3,-0.1,0.6
file3.csv:
a,b,c,d
-0.5,0.6,0.5,0.9
-0.8,0.7,-0.3,0.8
...
-0.9,-0.8,-0.3,0.3
Given three example files which can be used for training and evolving the networks (either RNNs or GPs) as well as validating their results to calculate the fitness. These are a four column CSV files with the first column being named a, the second column being named b and so on. These column names can be used to specifiy which columns are used as inputs to the evolved networks. The files used for training are specified with the --training_filenames <str>+ command line option and the files used for validation are specified with the --validation_filenames <str>+ command line option. Similarly, the --input_parameter_names <str>+ specify which columns are used as inputs to the networks and --output_parameter_names <str>+ specify which columns are being predicted (i.e., the outputs of the networks). Note that the same columns can be used for both inputs and outputs.
As the networks evolved are used for time series forecasting, the --time_offset <int> command line option specifies how far in the future (how many rows) the network is predicting. So if --time_offset 5 is specified the values from row 1 would be used to predict the values in row 6, the values in row 2 would be used to predict the values in row 7, and so on. --time_offset can also be set to 0 to predict the input data, which can be useful for evolving auto-encoder like networks.
EXAAM and EXA-GP currently utilize unbatched stochastic gradient descent to train the evolved networks, so each training file specified is used as a sample which are randomly shuffled each epoch. We have found however that while memory cell recurrent architectures are supposed to well handle long term time dependencies in practice this is not necessarily the case. It is possible to improve performance by dividing up input time series data into smaller sequences7. The --train_sequence_length <int> command line option can be used to specify how many rows to slice each training file into (if they are not evenly divisible by this number the last slice will be the remaining rows of the file).
If the training and validation CSVs are not already normalized, they can be normalized with the optional --normalize <str> argument which can either be min_max which will calculate the min and max value for each column in the training data, and use those values to normalize the data:
Or can be avg_std_dev which does computes average and standard deviation of the training data columns and normalizes the data (i.e., z-score normalization):
Putting this all together, given the following command line options and the above example files, we can run the multithreaded version of EXAMM (with ... being other options described in the upcoming section):
./multithreaded/examm_mt --training_filenames file1.csv file2.csv --validation_filenames file3.csv --input_parameter_names a b d --output_parameter_names c d --time_offset 1 --train_sequence_length 50 --normalize avg_std_dev ...
Note that the min/max or avg/std dev values from the training data are used to normalize the validation data.
This will run EXAMM with file1.csv and file2.csv, each split up into segments of at most 50 rows, to train the evolved networks and calculate the fitness of those networks using file3.csv. Each file will be z-score normalized based on the training files. The values in columns a, b and d will be used to predict the values in columns c and d in the next row (a time offset of 1).
Given the above options for loading and using training and validation data, we can explore the various options for running EXAMM and EXA-GP. The library also contains an implementation which utilizes NEAT speciation, for comparison purposes, which also serves as a memetic (backprop enabled) version of NEAT8 with the advanced node level mutation operations of EXAMM.
The following command line options control the neuroevolution search process itself.
-
--max_genomes <int>specifies how many genomes (RNNs or GPs) to evaluate before terminating the run. Note that EXAMM/EXA-GP use an asynchronous strategy with steady state populations so there are no explicit generations. -
--min_recurrent_depth <int>and--max_recurrent_depth <int>specify the possible range of time skip values for recurrent connections added to the evolved networks. Default values are 1 and 10. Adding in deeper recurrent connections has been shown to improve forecasting performance, and in some cases even outperform memory cells2. -
--possible_node_types <str>+specifies the options for selecting which node types can be added to networks during the evolution process. Default possible node types are the default for EXAMM (simple,jordan,elman,ugrnn,mgu,gru,delta, andlstm(please see 1 for more details on these node types). EXA-GP can be enabled by instead usingsigmoid,tanh,sum,multiply,inverse,sinandcosas possible node types; and the better peforming EXA-GP-MIN can be enabled with the_gpoptions:sigmoid_gp,tanh_gp,sum_gp,multiply_gp,inverse_gp,sin_gpandcos_gpfor the possible node types (for more details on their implementation see 45). -
--speciation_method <str>specifies if genomes in the population should be speciated into islands (usingisland) or with NEAT's speciation strategy (usingneat). Each of these come with their own set of parameters (see subsections below):
--number_islands <int>specifies how many islands should be used to perform the search, with a minimum of 1. If only 1 island is specified this operates the same as a single population version.--island_size <int>specifies the maximum number of genomes each island will hold for its population.--extinction_event_generation_number <int>specifies how frequently to perform island extinction if the value (N) is greater than 0. After every N inserted genomesislands_to_exterminateislands selected byisland_ranking_methodwill have their genomes removed and these will be repopulated as specfied by therepopulation_method. See 6 for full details and an examination of this methodology.--islands_to_exterminate <int>specifies how many islands to repopulate in an extinction event.--island_ranking_method <str>currently only allowsEraseWorsewhich will have extinction happen on the island(s) with the lowest fitness of the island's best individual.--repopulation_method <str>allows forbestparents,randomparents,bestgenomeandbestisland:bestparentsselects 2 parents randomly from the best parents of other (non-repopulating) islands to perform crossover on to generate new genomes to repopulate islands.randomparentsselects 2 parents randomly from the genomes of all other non-repopulating islands to perform crossover on to generate new genomes to repopulate islands.bestgenomeselects the global best genome and performs mutations on it to repopulate islands.bestislandselects the best island and repopulates islands by performing a mutation on each genome in the best island.
--num_mutations <int>specifies how many mutation operations to perform when generating a child genome by mutation.--repeat_extinctionif not specified, if an island is repopulated it will not be repopulated until 5 other extinction events have passed. This prevents the same island from being repopulated over and over. Turning this flag on allows islands to be repeatedly repopulated.
If neat is selected as the speciation method, the following hyperparameters from the NEAT paper8 can be specified.
--species_threshold <float>--fitness_threshold <float>--neat_c1 <float>--neat_c2 <float>--neat_c3 <float>
Given the following equation, where neat_c1 is the neat_c2 is the neat_c3 is the
If species_threshold,
Where fitness_threshold. Using the above hyperparameters genomes will be placed into species as done in the NEAT algorithm.
EXAMM allows for genomes to be initialized using uniform random, Xavier, Kaiming or a Lamarckian weight inheritance strategy, as described in3. For EXA-GP-MIN there is also a genetic programming weight initialization, where edge weights are fixed to 1, and the initial network has weights all set to 0 except for weights going from an input node to the output node for the same parameter, as per5. It allows for different initialization methods to be used at different points in the search strategy. Options for each are random (uniform random), xavier, kaiming, lamarckian, or gp (for EXA-GP-MIN).
--weight_initialize <str>specifies how weights are generated in the initialization phase of the EXAMM algorithm while the initial genomes are being generated (without parents).--weight_inheritance <str>specifies how weights are inherited from parent genomes after the initialization phase of EXAMM. Lamarckian inheritance allows the reuse of parental weights, while other methods will re-initialize the genome from scratch.--mutated_component_weight <str>specifies how new weights are generated on a mutation (e.g., ifweight_inheritanceis set tolamarckianthe genome will reuse its parental weights, and then weights for new components generated by mutation could be generated randomly withrandom,xavierorkaiming; or using the parental weight distribution withlamarckian`).
The following allow control of the neural network training hyperparameters:
--bp_iterations <int>specifies how many backpropagation epochs should be done per genome.
-
--learning_rate <float>specifies the learning rate,$\alpha$ , (which will be utilized by the varyingweight_updateoptimizer options below). -
--high_threshold <float>(default 1.0) specifies the threshold used for gradient scaling (to help prevent exploding gradients), as presented by Pascanu et al.9, where the gradient calculated by backpropagation,$g$ , is reduced if the L2 norm of the gradient is above a threshold,$t_{high}$ :
-
--low_threshold <float>(default 0.005) performs gradient boosting (the opposite of gradient scaling), to help with vanishing gradients. This is unpublished work but we have found it improves training performance. When the L2 norm of a gradient is below a threshold,$t$ , the gradient is increased if the L2 norm of the gradient is below a threshold,$t_{low}$ :
Using --weight_update <str> specifies the optimizer used for performing weight updates, with
vanillaperforms a vanilla weight update:
-
momentumperforms a weight update with momentum, given$\mu$ as--mu <float>(default 0.9):
-
nesterovperforms a weight update using Nesterov momentum, given$\mu$ as--mu <float>(default 0.9):
-
adagradperforms Adagrad with, given$\epsilon$ as--eps <float>(default 1e-8):
-
rmspropperforms RMSProp using$\epsilon$ as--eps <float>(default 1e-8) and$\gamma$ as--decay_rate <float>(default 0.9):
-
adamperforms Adam (without bias correction) given$\epsilon$ as--eps <float>(default 1e-8),$\beta_1$ as--beta1 <float>(default 0.9), and$\beta_2$ as--beta2 <float>(default 0.99):
-
adam-biasperforms full Adam with bias correction given$\epsilon$ as--eps <float>(default 1e-8),$\beta_1$ as--beta1 <float>(default 0.9), and$\beta_2$ as--beta2 <float>(default 0.99):
EXAMM provides a number of options for tracking results of its neuroevolution runs. The following provide options for saving generated neural networks in a number of ways, as well as where to put various log files for analysis. Neural networks or genetic programs will be saved with three or four files (.txt, .gv, .bin and optionally .json, see below).
The .bin file is a serialized binary of the network, the .txt file is a textual representation of the network or genetic program equations, and the .gv file is a graphviz file of the network so a visualization of the network can be created with graphviz (if graphviz is installed you can run dot -T pdf <file>.gv -o <file>.pdf to create a PDF representation of the network or genetic program. The .json file is a JSON representation of the network so it can be utilized in other applications, such as the Genetic Distance Projection visualization framework10.
-
--output_directory <str>specifies a directory where all output files (log files and neural network files) will be placed. This directory will be created it if it does not exist. This directory will contain afitness_log.csvfile which tracks inforomation for every genome inserted into the population, including time, fitness, per-island fitness and some network statistics. When the search completes it will also contain an emptycompletedfile if the search completed without error. It will also containglobal_best_genome_<generation_id>files (bin, txt and gv) for the global best neural network (or genetic program) found.generation_idis a counter specifing the order in which genomes were generated (not inserted) by the search. -
--save_genome_option <str>(default =all_best_genomes) specifies which generated genomes to save. Currently the only option isall_best_genomeswhich will save the.gv,.binand.txtfile of every new global best genome found in the specifiedoutput_directoryasrnn_genome_<generation_id>.<extension>. -
--generate_visualization_jsonif this flag is specified (default false), every evaluated genome will generate arnn_genome_<generation_id>.jsonfile so that the entire neuroevolution run can be visualized using the Genetic Distance Projection visualization framework, or loaded by other applications for analysis. The JSON file contains all nodes, connections and weights of the network or genetic program. -
--generate_op_logif this flag is specified (default false), an additional operation log file will be generated which will track for every genome how it was generated (i.e., which mutation or crossover operation(s) were used) as well has how many nodes of what type (e.g., LSTM, GRU, MGU, etc) were generated and used. This is turned off by default as generating this log file is somewhat slow and can degrade performance.
As discussed in the previous section, EXAMM will save the best found generated networks for its neuroevolution runs in binary .bin files. These are serialized so that their results are reproducible from the neuroevolution run. Please note that, networks generated from JSONs may not provide the exact same results due to conversion from double precision weights to text and back. There is one file in particular that is useful for utilizing these evolved neural networks and genetic programs:
- evaluate_rnn.cxx can take a target set of testing files (in the expected format from datasets above) which will evaluate the input RNN (specified by the
--genome_file <file.bin>command line argument) on the testing files (specified by--testing_filenames <str>+). Note that the genome binary files save the normalization values and methodology used on the training data so that these same normalization values are used on the testing data (this methodology does not cheat by utilizing normalization statistics from potentially unknown test data). This will write files with the predictions of the RNN or genetic program to the specified--output_directory <str>with the output predicition filenames as<input_test_file>_predictions.csv.
EXACT (Evolutionary eXploration of Augmenting Convolutional Topologies) was a predecessor project focused on evolving convolutional neural networks. While the source code and documentation for EXACT is still available in this repository, setting it up requires specific configurations and dependencies. If you're interested in using EXACT, please contact us for instruction on setup and implementation. We're happy to help you get started with the system.
© 2025 Distributed Data Science Systems Lab (DS2L), Rochester Institute of Technology. All Rights Reserved.
Footnotes
-
Alex Ororbia, AbdElRahman ElSaid, and Travis Desell. Investigating Recurrent Neural Network Memory Structures using Neuro-Evolution. The Genetic and Evolutionary Computation Conference (GECCO 2019). Prague, Czech Republic. July 8-12, 2019. ↩ ↩2
-
Travis Desell, AbdElRahman ElSaid and Alexander G. Ororbia. An Empirical Exploration of Deep Recurrent Connections Using Neuro-Evolution. The 23nd International Conference on the Applications of Evolutionary Computation (EvoStar: EvoApps 2020). Seville, Spain. April 15-17, 2020. Best paper nominee. ↩ ↩2
-
Zimeng Lyu, AbdElRahman ElSaid, Joshua Karns, Mohamed Mkaouer, Travis Desell. An Experimental Study of Weight Initialization and Lamarckian Inheritance on Neuroevolution. The 24th International Conference on the Applications of Evolutionary Computation (EvoStar: EvoApps 2021). ↩ ↩2
-
Jared Murphy, Devroop Kar, Joshua Karns, and Travis Desell. EXA-GP: Unifying Graph-Based Genetic Programming and Neuroevolution for Explainable Time Series Forecasting. Proceedings of the Genetic and Evolutionary Computation Conference Companion. Melbourne, Australia. July 14-18, 2024. ↩ ↩2
-
Jared Murphy, Travis Desell. Minimizing the EXA-GP Graph-Based Genetic Programming Algorithm for Interpretable Time Series Forecasting. Proceedings of the Genetic and Evolutionary Computation Conference Companion. Melbourne, Australia. July 14-18, 2024. ↩ ↩2 ↩3
-
Zimeng Lyu, Joshua Karns, AbdElRahman ElSaid, Mohamed Mkaouer, Travis Desell. Improving Distributed Neuroevolution Using Island Extinction and Repopulation. The 24th International Conference on the Applications of Evolutionary Computation (EvoStar: EvoApps 2021). ↩ ↩2
-
Zimeng Lyu, Shuchita Patwardhan, David Stadem, James Langfeld, Steve Benson, and Travis Desell. Neuroevolution of Recurrent Neural Networks for Time Series Forecasting of Coal-Fired Power Plant Data. ACM Workshop on NeuroEvolution@Work (NEWK@Work}, held in conjunction with ACM Genetic and Evolutionary Computation Conference (GECCO). pp. 1735-1743. Lille, France. July 10-14, 2021. ↩
-
Kenneth Stanley and Risto Miikkulainen. Evolving neural networks through augmenting topologies. Evolutionary Computation 10.2(2002): 99-127. ↩ ↩2
-
Razvan Pascanu, Tomas Mikolov and Yoshio Bengio. On the Difficulty of Training Recurrent Neural Networks. The International Conference on Machine Learning (ICML 2013). 2013. ↩
-
Evan Patterson, Joshua Karns, Zimeng Lyu and Travis Desell. Visualizing the Dynamics of Neuroevolution with Genetic Distance Projections. The Genetic and Evolutionary Computation Conference (GECCO 2025). Malaga, Spain. July 2025. ↩