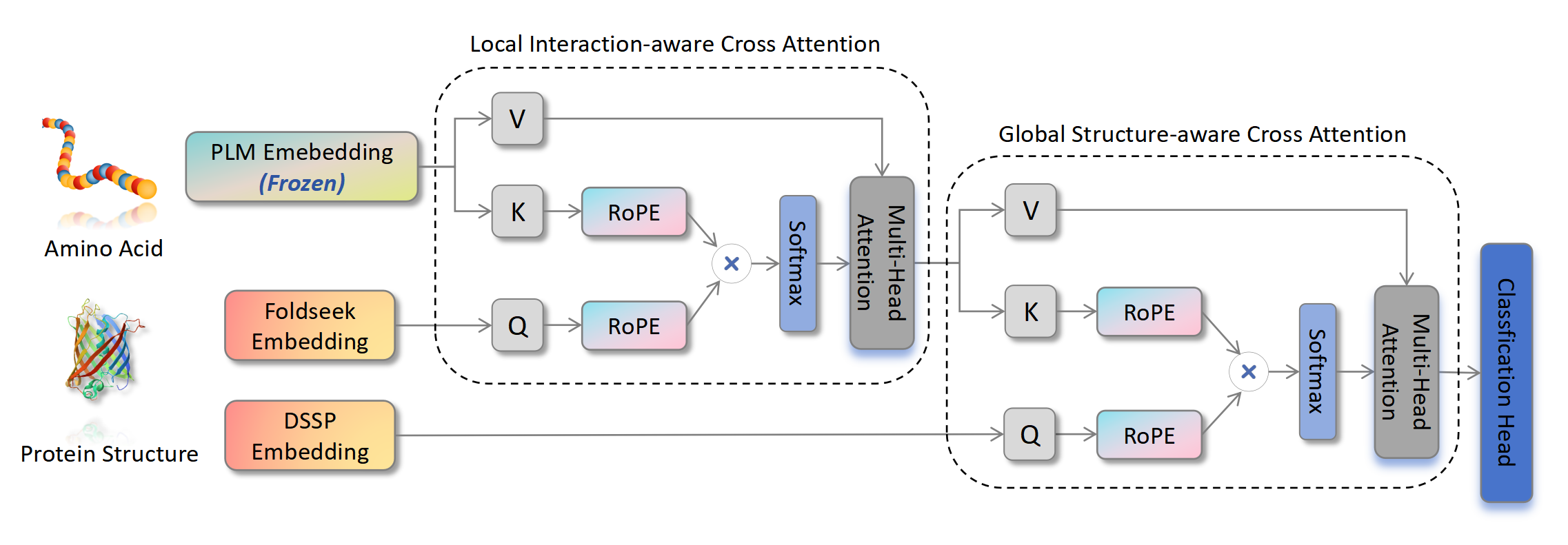
SES-Adapter, a simple, efficient, and scalable adapter method for enhancing the representation learning of protein language models (PLMs).
We serialized the protein structure and performed cross-modal-attention with PLM embeddings, effectively improving downstream task performance and convergence efficiency.
- [2024.07.31] Congratulations! Our paper was accepted by Journal of Chemical Information and Modeling!
- [2024.07.08] We have added the ESM3 structure tokenizer in
data/get_esm3_structure_seq.py, you should download the checkpoint from ESM3 huggingface andpip install esm.
We conduct evaluation on 9 state-of-the-art baseline models (ESM2, ProtBert, ProtT5, Ankh) across 9 datasets under 4 tasks (Localization, Function, Solubility, Annotation).
Results show that compared to vanilla PLMs, SES-Adapter improves downstream task performance by a maximum of 11% and an average of 3%, with significantly accelerated training speed by a maximum of 1034% and an average of 362%, the convergence rate is also improved by approximately 2 times.
Please make sure you have installed Anaconda3 or Miniconda3.
conda env create -f environment.yaml
conda activate ses_adapter
We recommend a 24GB RTX 3090 or better, but it mainly depends on which PLM you choose.
We provide datasets and format references in the dataset folder. We support both JSON and CSV data formats.
- JSON:
- example: https://github.com/tyang816/SES-Adapter/tree/main/dataset/BP
- For more JSON dataset, you can found in dataset.
- CSV example:
- example: https://huggingface.co/datasets/tyang816/DeepLocBinary_AlphaFold2
- For more CSV dataset, you can found in huggingface.
Config file should be specified ahead.
{
"dataset": "BP", # Huggingface: tyang816/BP_AlphaFold2
"pdb_type": "AlphaFold2",
"train_file": "dataset/BP/AlphaFold2/train.json", # no need for Huggingface
"valid_file": "dataset/BP/AlphaFold2/valid.json", # no need for Huggingface
"test_file": "dataset/BP/AlphaFold2/test.json", # no need for Huggingface
"num_labels": 1943,
"problem_type": "multi_label_classification",
"metrics": "f1_max", # for multiple metrics use ',' to split, example: "accuracy,recall,precision"
"monitor": "f1_max",
"normalize": "None"
}
| Metric Name | Full Name | Problem Type |
|---|---|---|
| accuracy | Accuracy | single_label_classification/ multi_label_classification |
| recall | Recall | single_label_classification/ multi_label_classification |
| precision | Precision | single_label_classification/ multi_label_classification |
| f1 | F1Score | single_label_classification/ multi_label_classification |
| mcc | MatthewsCorrCoef | single_label_classification/ multi_label_classification |
| auc | AUROC | single_label_classification/ multi_label_classification |
| f1_max | F1ScoreMax | multi_label_classification |
| spearman_corr | SpearmanCorrCoef | regression |
See the train.py script for training details. Examples can be found in scripts folder.
Please cite our work if you have used our code or data.
@article{tan2024ses-adapter,
title={Simple, Efficient, and Scalable Structure-Aware Adapter Boosts Protein Language Models},
author={Tan, Yang and Li, Mingchen and Zhou, Bingxin and Zhong, Bozitao and Zheng, Lirong and Tan, Pan and Zhou, Ziyi and Yu, Huiqun and Fan, Guisheng and Hong, Liang},
journal={Journal of Chemical Information and Modeling},
year={2024},
publisher={ACS Publications}
}