We introduce a suite of neural language model tools for pre-training, fine-tuning SMILES-based molecular language models. Furthermore, we also provide recipes for semi-supervised recipes for fine-tuning these languages in low-data settings using Semi-supervised learning.
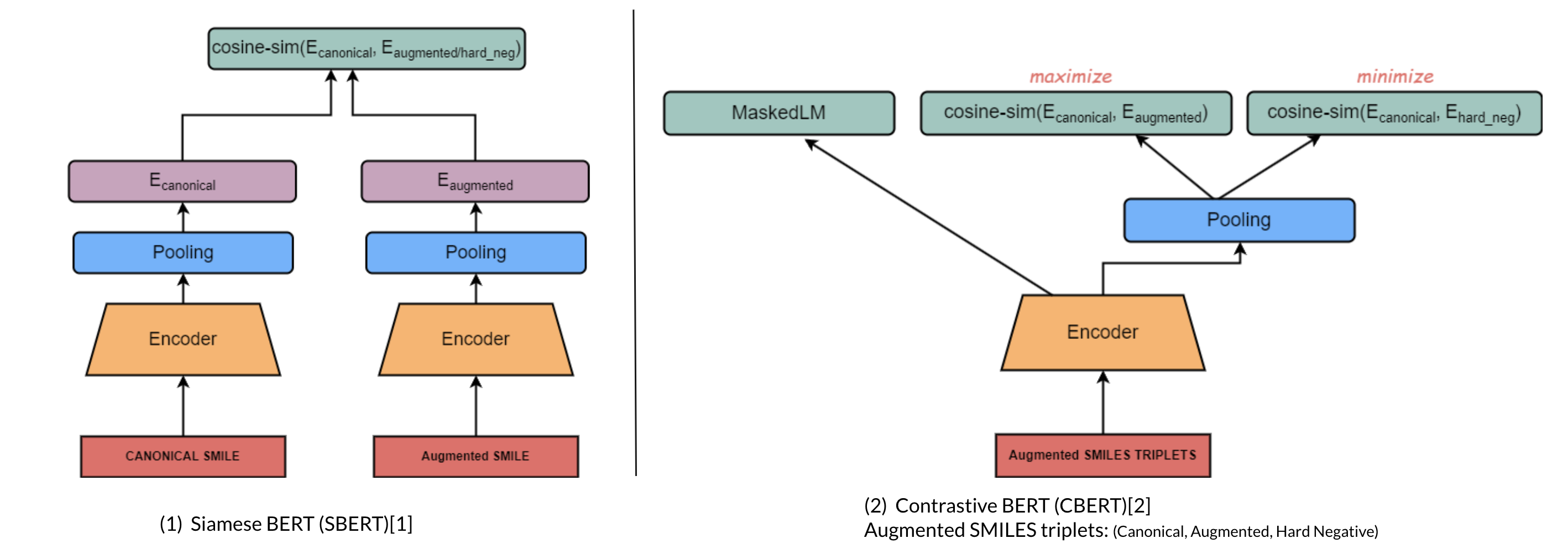
Introduces contrastive learning alongside multi-task regression, and masked language modelling as pre-training objectives to inject enumeration knowledge into pre-trained language models.
You can reproduce the experiment by:
bash pip install -r requirements.txt
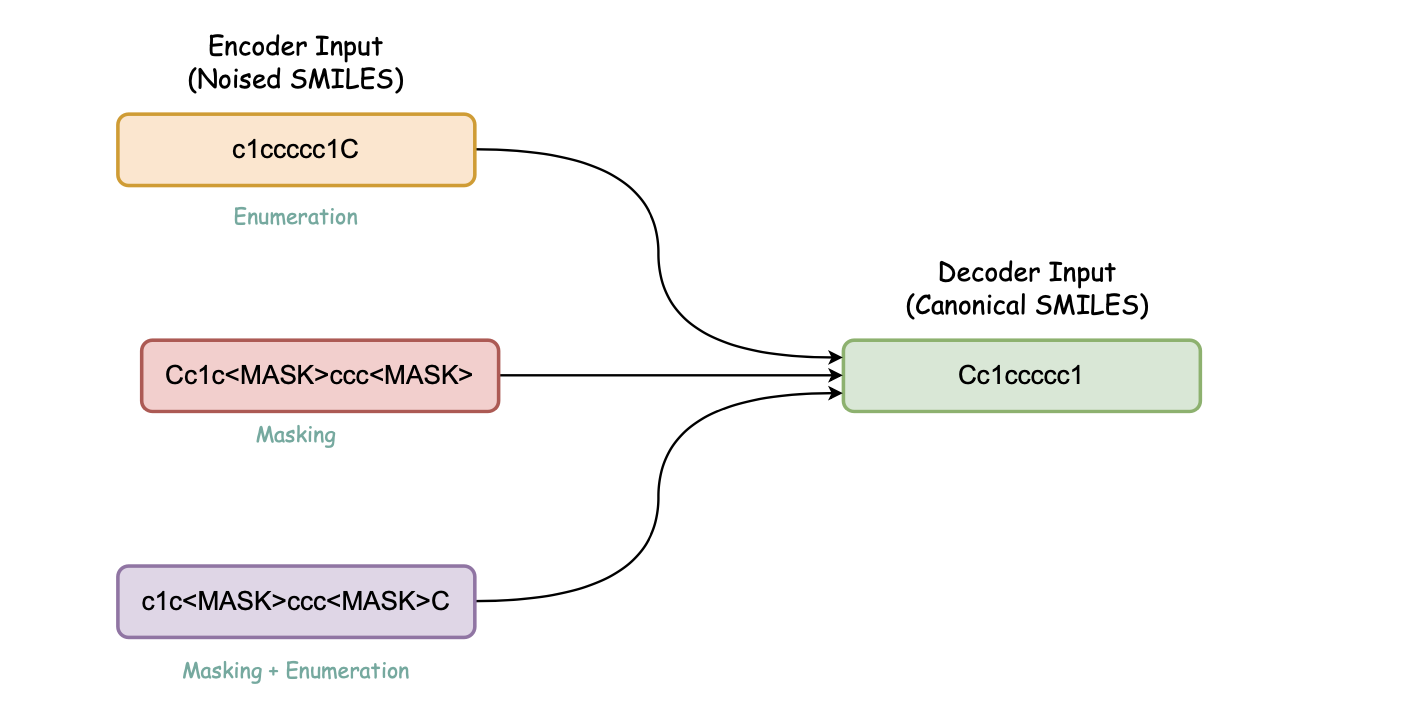
The detailed steps are for pre-training with Encoder-based architectures pertaining MLM, MTR and Seq2Seq BART with denoising objectives are outlined in here.
To reproduce the domain adaptation step from our work please follow the guidelines here.
Finally for finetuning the domain adapted molecular languages on downstream tasks are explained in the accompanying notebook which can be found here.
Code base adapted from: