Python3 scripts for string dataset generator for clustering purpose.
It is important to note thatThis implementation relies heavily on Jaccard's similarity method.
mkdir datamake sure you have your data file in there.
Let's say you have a data file which contains a list of patients with icd-10 diseases.
The data file will look like the following
L01 K11 R44 X09 F00
F00 R21
M12 B20 L40 K50
.
.
.
W08 Q90 P00
# this will execute the example from main
$ cd src/
$ python3 main.pyTo use this program as a library you can simple import artificial_set_data_generator in to your python module the call the following function artificial_set_data_generator.generate.
- data_size : (int) an integer number specifies number of total number of data that will be generated
- size_of_clusters : (numpy arry) specifies size for each cluster. If empty array is passed then the size of all cluster will be the same. Note that len of array should equal to number_of_cluster and sum of this array should equal to data_size
- number_of_cluster : (int) an integer number specifies number of cluster to create
- dimension : (int) an integer number specifies total number of features that will be generate in the data set
- distance_threshold : (float) a number specifies the maximum distance away from the cluster representative according to Jaccard's method
- size_of_set : (tuple(int,int)) a tuple of intergers specifies the minimum and maximum feature that each data has to contain
- all_features : (string[]) an array of string containing all possible features of the dataset
- gt_representative : (string[][]) if empty the program will randomly generate the cluster representatives, else program uses the provides values as cluster representatives
As mentioned above, you can do so by given them through gt_representative
If you run the code from our main.py you can store your ground truth representatives in data directory and change the path in GT_REPRESENTATIVE_FILE_PATH accordingly.
Make sure that your stored representatives are store in the same format as the original input (take a look at the example above).
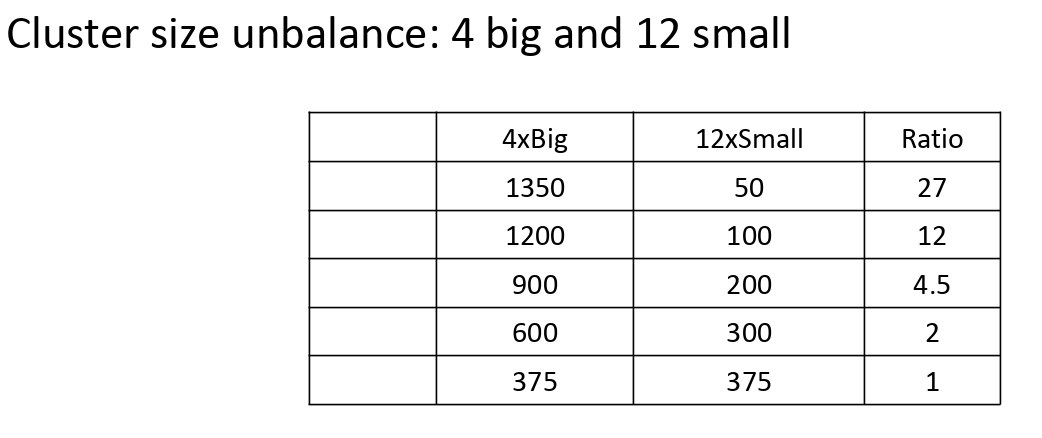
From the configuration section, there is an option that you can provide different sizes for each cluster (array with integers instead of empty for size_of_clusters)
The library provides some APIs to help generate those values in the pattern that have been found common.
Those APIs located in size.py under imbalance directory
-
random_cluster_sizes :
- arguments: (int) data size, (int) number of cluster
- output: (int[]) integers array with len of number of cluster, each element will be a random number that in total will sum to the amount of data size.
-
build_specific_sizes :
- arguments (int) data size, ((int, int)) a tuple of 2 integers specify number of big and small clusters accordingly, (int) ratio of big and small cluster member
- output: (int[]) integers array with len of number of cluster with specified number of big and small clusters. The value of each element will be the number of cluster according to the given ratio.
From the main.py example, the output will be written the 3 separate files in out folder.
- gen_data{}N_{}K_{}overlap.txt
- gt_representative{}N_{}K_{}overlap.txt
- gt_labels{}N_{}K_{}overlap.txt
note: {} before N will be formatted with number of generated data you specified
note2: {} before K will be formatted with number of generated cluster you specified
note3: {} before overlap stand for the percentage of data overlap that program generated
- improving control over overlap percentage. Ideally, users should be able to specify the overlap percentage as on amoung other program's configurations