03.31.2021 we find some docs are broken and some are out-of-date, so we do a major clean on docs. Please refer to the latest doc. We strongly recommend picking up a demo example from our demo list*, and modify your own task based on it.
The EasyReg is a image processing package, that supports a series of mainstream methods.
We list a few properties:
- A simple interface for learning/optimization based image registration tools, e.g., Mermaid and other popular registration packages*.
- A simple interface for a series of data augmentation approaches, supporting both random data augmentation and anatomical data augmentation/inter-and extra-polation
- A simple interface for image segmentation.
*The currently supported methods include Mermaid-optimization (i.e., optimization-based fluid registration) and Mermaid-network (i.e., deep network-based fluid registration methods using the mermaid deformation models). We also added support for ANTsPy, NiftyReg, Demons(embedded in SimpleITK), VoxelMorph and its diffeomorphic variant, though we recommend using the official release.
Currently, we support 3d image registration (2d is in progress).
The latest doc can be found here https://easyreg-unc-biag.readthedocs.io/en/latest/.
Source code documentation and tutorials can be built on local using sphinx.
cd EASYREG_REPOSITORY_PATH
cd doc
make html
Now you are ready to explore various optimization-based as well as learning-based demos provided by EasyReg.
We provide a series of demos for both learning and optimization methods :)
Demo list (for more details, please refer to the doc*)
- ANTsPy on OAI (knee MRI of the Osteoarthritis Initiative dataset)
- NiftyReg on OAI
- Demons on OAI
- Optimization-based mermaid registration on OAI (vSVF)
- Optimization-based mermaid registration on lung pairs** (inspiration to expiration) (RDMM)
- Pretrained learning-based mermaid registration on OAI (vSVF, RDMM)
- A training demo for joint affine and vSVF registration on sub-OAI dataset (3 pairs)
- A training demo for VoxelMorph like registration framework (cvpr and miccai version)
* For 2D demo (vSVF, LDDMM, RDMM) on synthetic data, please refers to mermaid
** Thanks Dr. Raúl San José Estépar for providing the lung data
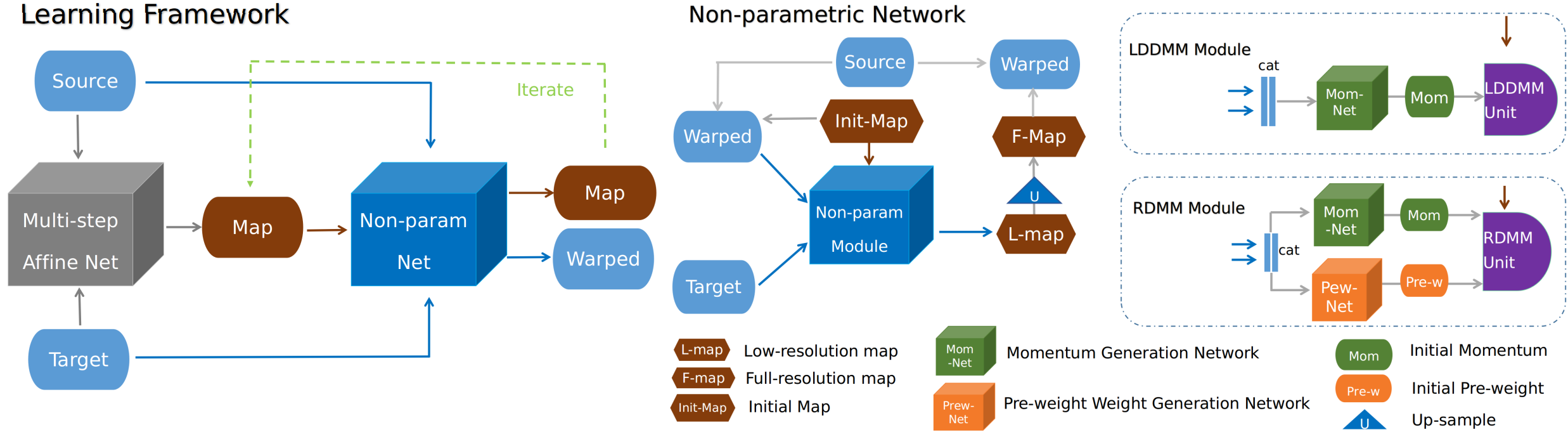
For the learning part, the easyreg provides a two-stage learning framework including affine registration and non-parametric registration (map-based).
An illustration of learning architecture
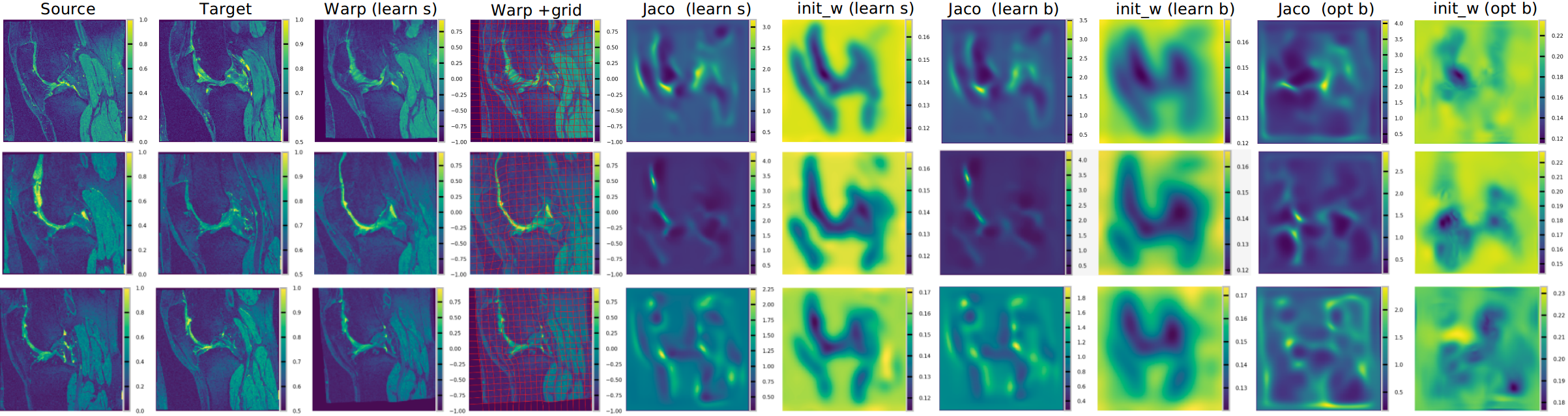
Registration results from Region-spec Region-specific Diffeomorphic Metric Mapping (RDMM)
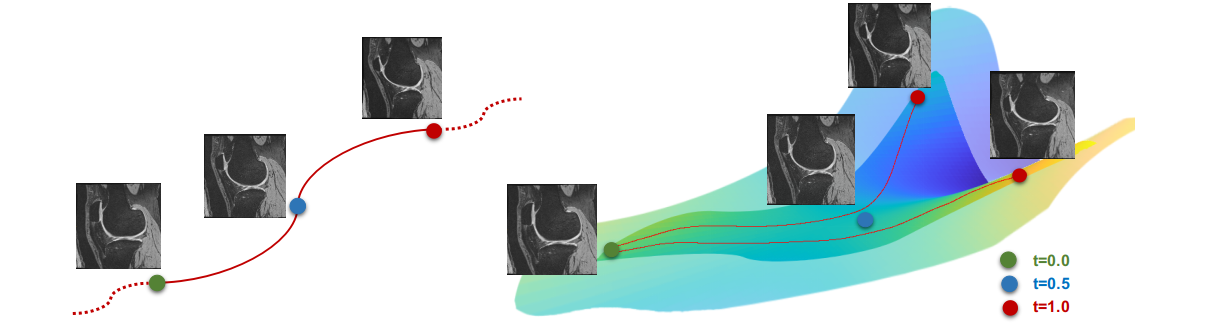
We provide an easy interface for fluid-based data augmentation and interpolation/extrapolation. A typical strategy is as following:
- Construct a geodesic subspace with given a source image and a set of target images.
- Sample transformations from the resulting geodesic subspace.
- Obtain deformed images and segmentations via interpolation.
The current supported method list:
- Anatomical fluid-based data augmentation
- Random data augmentation (including Bspline random transformation and Random momentum based fluid data augmentation)
- Brainstorm like data-augmentation framework
Related demos can be found at Demos on Data augmentation section.
An illustration of sampling from geodesic subspace.
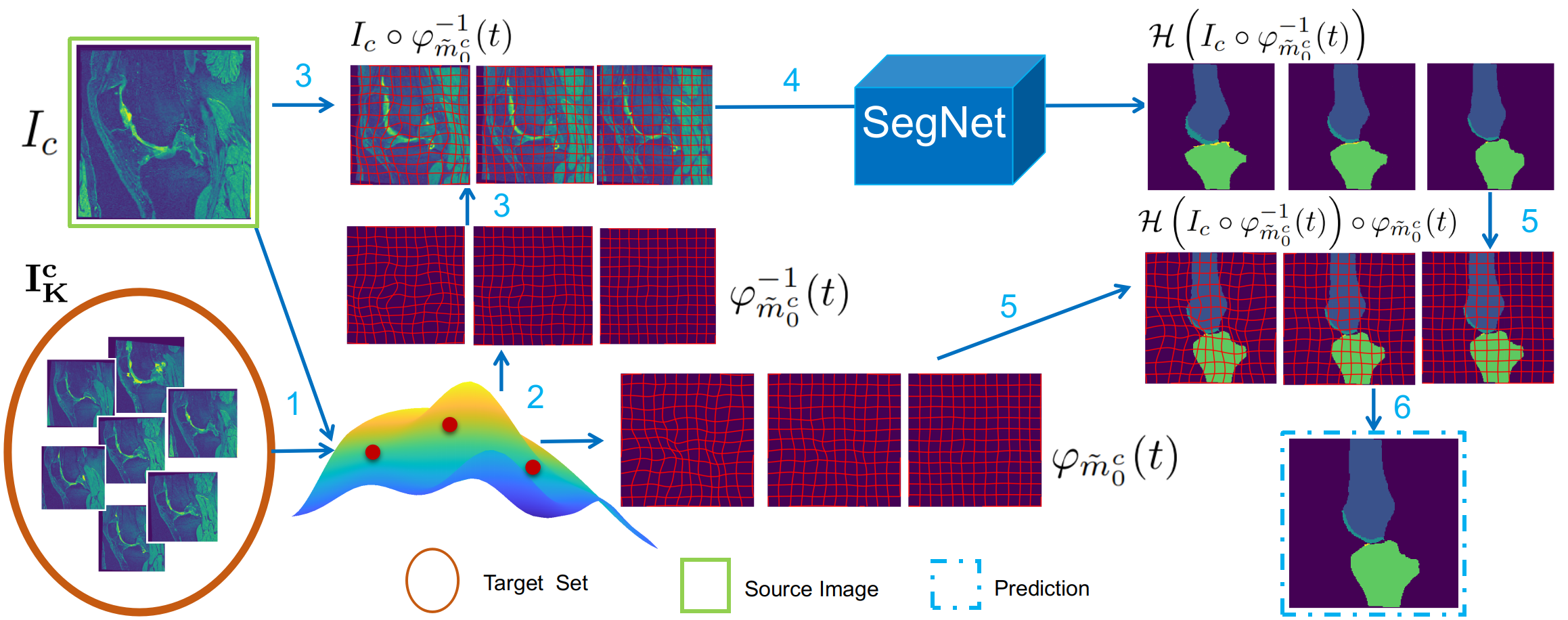
We provide an interface for image segmentaion task with a u-net structure. The segmentation framework supports an test phase data augmentation method that can generally improve the performance without additional models.
An illustration of data augmentation- based ensemble prediction during test.
conda create -n easyreg python=3.6
conda activate easyreg
git clone https://github.com/uncbiag/easyreg.git
cd easyreg
git clone https://github.com/uncbiag/mermaid.git
pip install -r requirements.txt
# ################download demo (optional)######
cd demo
gdown https://drive.google.com/uc?id=1yAqks0KEK9pYMKc1W-1714cwN_PONrYU
unzip demo.zip -d .
cd ..
# #############################################
cd mermaid
python setup.py develop
For third-party toolkits:
ANTsPy
We currently test using ANTsPy version 0.1.4. Since AntsPy is not fully functioned, it will be replaced with a custom Ants Package in the next release. The following command installs ANTsPy 0.1.4.
pip install https://github.com/ANTsX/ANTsPy/releases/download/v0.1.4/antspy-0.1.4-cp36-cp36m-linux_x86_64.whl
NiftyReg
For NiftyReg installation instructions please refer to NiftyReg Installation
If you find EasyReg is helpful, please cite (see bibtex):
Anatomical Data Augmentation via Fluid-based Image Registration [link]
Zhengyang Shen, Zhenlin Xu, Sahin Olut, Marc Niethammer. MICCAI 2020.
Region-specific Diffeomorphic Metric Mapping [link]
Zhengyang Shen, François-Xavier Vialard, Marc Niethammer. NeurIPS 2019.
Networks for Joint Affine and Non-parametric Image Registration [link]
Zhengyang Shen, Xu Han, Zhenlin Xu, Marc Niethammer. CVPR 2019.
See https://github.com/uncbiag/registration for an overview of other registration approaches of our group and a short summary of how the approaches relate.