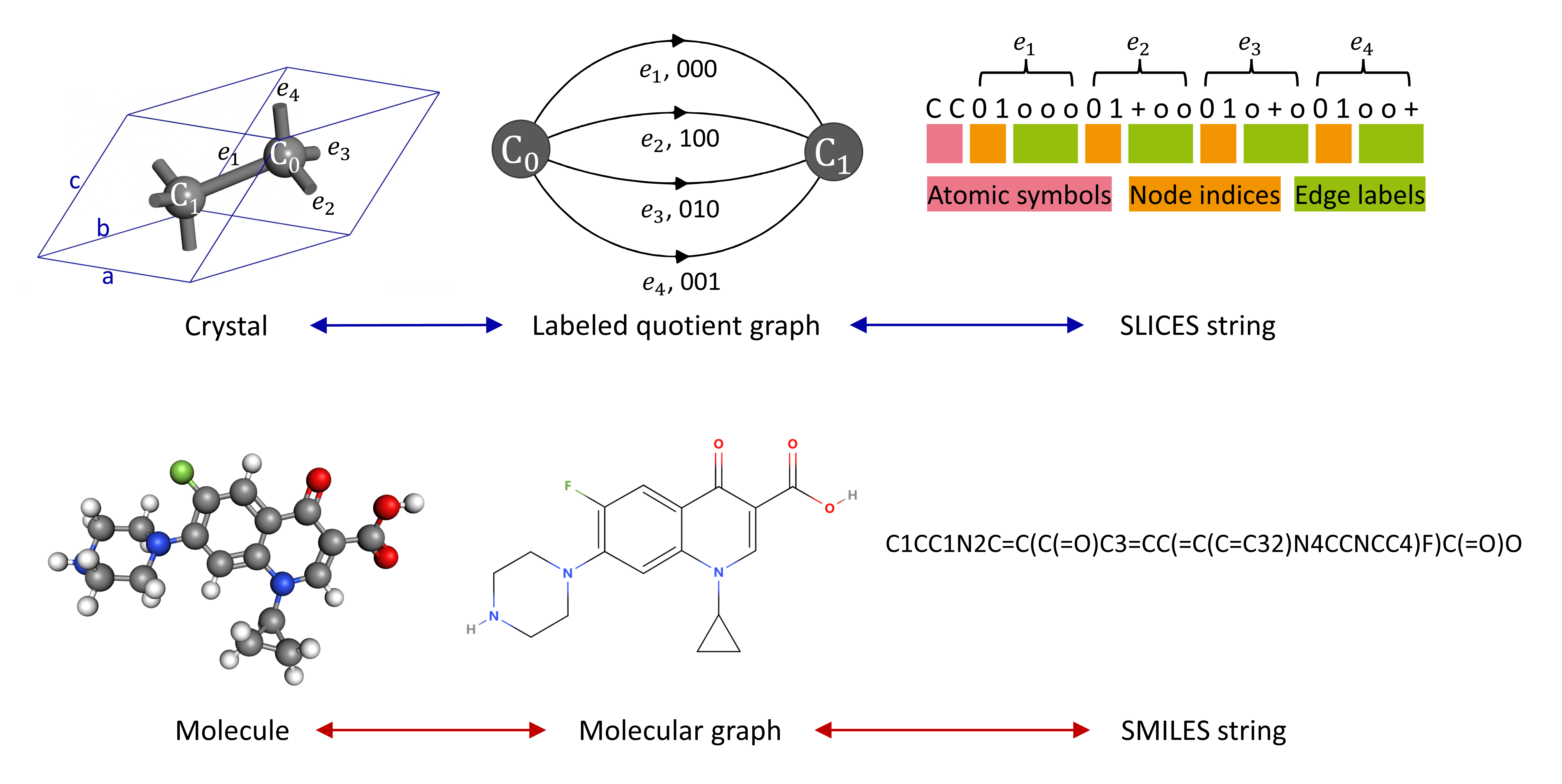
Text2Crystal. Simplified line-input crystal-encoding system (SLICES): An Invertible, Symmetry-invariant, and String-based Crystallographic Representation
Scripts to reproduce results of benchmark and the inverse design case study in https://chemrxiv.org/engage/chemrxiv/article-details/64697e40a32ceeff2df995c0.
Developed by Hang Xiao 2023.04 xiaohang007@gmail.com https://github.com/xiaohang007
Training datasets and results of the inverse design study are available in https://doi.org/10.6084/m9.figshare.22707472.
The source code of SLICES is in ./invcryrep/invcryrep/invcryrep.py and ./invcryrep/invcryrep/tobascco_net.py.
!!! We also provide a codeocean capsule (a modular container for the software environment along with code and data, that runs in a browser), allowing one-click access to guaranteed computational reproducibility of SLICES's benchmark. https://codeocean.com/capsule/8643173/tree/v1
Download this repo and unzipped it.
Put Materials Project's new API key in "APIKEY.ini".
Edit "CPUs" in "slurm.conf" to set up the number of CPU threads available for the docker container.
It is recommemded to run this docker image under Linux. Running on docker on windows using WSL2.0 is possible, but the inverse transform could be stuck in an uninterruptible sleep (D) state due to a weird bug of running m3gnet in docker container using WSL2.
docker pull xiaohang07/slices:v2 # Download SLICES_docker with pre-installed SLICES and other relevant packages.
# Repalce "[]" with the absolute path of this repo's unzipped folder to setup share folder for the docker container.
docker run -it -h workq --shm-size=0.1gb -v /[]:/crystal -w /crystal xiaohang07/slices:v2 /crystal/entrypoint_set_cpus.sh !!! In case "docker pull xiaohang07/slices:v2" is not working, one can download SLICES_docker.tar.gz.* from https://doi.org/10.6084/m9.figshare.22707946 with pre-installed SLICES and other relevant packages.
cat SLICES_docker.tar.gz.* | tar xzvf - # Uncompress SLICES_docker.tar.gz.*
docker load -i SLICES_docker_image.tar.gz # Install the docker image
# Repalce "[]" with the absolute path of this repo's unzipped folder to setup share folder for the docker container.
docker run -it -h workq --shm-size=0.1gb -v /[]:/crystal -w /crystal crystal:80 /crystal/entrypoint_set_cpus.sh2. Introductory example: converting a crystal structure to its SLICES string and converting this SLICES string back to its original crystal structure.
Suppose we wish to convert the crystal structure of NdSiRu (mp-5239,https://next-gen.materialsproject.org/materials/mp-5239?material_ids=mp-5239) to its SLICES string and converting this SLICES string back to its original crystal structure. The python code below accomplishes this:
import os
from invcryrep.invcryrep import InvCryRep
from pymatgen.core.structure import Structure
# setup modified XTB's path
os.environ["XTB_MOD_PATH"] = "/crystal/xtb_noring_nooutput_nostdout_noCN"
# obtaining the pymatgen Structure instance of NdSiRu
original_structure = Structure.from_file(filename='NdSiRu.cif')
# creating an instance of the InvCryRep Class (initialization)
backend=InvCryRep()
# converting a crystal structure to its SLICES string
slices_NdSiRu=backend.structure2SLICES(original_structure)
# converting a SLICES string back to its original crystal structure and obtaining its M3GNet_IAP-predicted energy_per_atom
reconstructed_structure,final_energy_per_atom_IAP = backend.SLICES2structure(slices_NdSiRu)
print('SLICES string of NdSiRu is: ',slices_NdSiRu)
print('\nReconstructed_structure is: ',reconstructed_structure)
print('\nfinal_energy_per_atom_IAP is: ',final_energy_per_atom_IAP,' eV/atom')
# if final_energy_per_atom_IAP is 0, it means the M3GNet_IAP refinement failed, and the reconstructed_structure is the ZL*-optimized structure.Convert MP-20 dataset to json (cdvae/data/mp_20 at main · txie-93/cdvae. GitHub. https://github.com/txie-93/cdvae (accessed 2023-03-12))
cd /crystal/benchmark/0_get_mp20_json
python 0_mp20.pyRule out unsupported elements
cd /crystal/benchmark/1_element_filter
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect.pyConvert to primitive cell
cd /crystal/benchmark/2_primitive_cell_conversion
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect.pyRule out crystals with low-dimensional units (e.g. molecular crystals or layered crystals)
cd /crystal/benchmark/3_3d_filter
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect.pyCalculate reconstruction rate of IAP-refined structures, L-optimized structures, rescaled structures under strict or coarse setting.
cd /crystal/benchmark/matchcheck3
python 1_ini.py # wait for jobs to finish (using qstat to check)
python 2_collect_grid_new.pyCalculate reconstruction rate of IAP-refined structures, L-optimized structures, IAP-refined rescaled structures, rescaled structures under strict or coarse setting.
cd /crystal/benchmark/matchcheck4
python 1_ini.py # wait for jobs to finish (using qstat to check)
python 2_collect_grid_new.pyDownload entries to build general and transfer datasets
cd /crystal/HTS/0_get_json_mp_api
python 0_prior_model_dataset.py
python 1_transfer_learning_dataset.py
!!! If “mp_api.client.core.client.MPRestError: REST query returned with error status code” occurs. The solution is:
pip install -U mp-apiRule out crystals with low-dimensional units (e.g. molecular crystals or layered crystals) in general dataset
cd /crystal/HTS/0_get_json_mp_api/2_filter_prior_3d
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect.pyRule out crystals with low-dimensional units (e.g. molecular crystals or layered crystals) in transfer dataset
cd /crystal/HTS/0_get_json_mp_api/2_filter_transfer_3d
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect.pyConvert crystal structures in datasets to SLICES strings and conduct data augmentation
cd /crystal/HTS/1_augmentation/prior
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect.py
cd /crystal/HTS/1_augmentation/transfer
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect.pyTrain general and specialized RNN; sample SLICES
cd /crystal/HTS/2_train_sample
sh 0_train_prior_model.sh
sh 1_transfer_learning.shModify /crystal/HTS/2_train_sample/workflow/2_sample_HTL_model_100x.py to define the number of SLICES to be sampled
sh 2_sample_in_parallel.sh # wait for jobs to finish (using qstat to check)
python 3_collect_clean_glob_details.pyReconstruct crystal structures from SLICES strings
cd /crystal/HTS/3_inverse
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.py
!!! In order to address the potential memory leaks associated with M3GNet, we implemented a strategy of
restarting the Python script at regular intervals, with a batch size of 30.Filter out crystals with compositions that exist in the Materials Project database
cd /crystal/HTS/4_composition_filter
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.pyFind high-symmetry structures in candidates with duplicate compositions
cd /crystal/HTS/5_symmetry_filter_refine
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.pyRule out crystals displaying minimum structural dissimilarity value < 0.75 (a dissimilarity threshold used in the Materials Project) with respect to the structures in the training dataset
cd /crystal/HTS/6_structure_dissimilarity_filter
cd ./0_save_structure_fingerprint
cp /crystal/HTS/0_get_json_mp_api/prior_model_dataset_filtered.json ./
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.py
cd ../
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.pyRule out candidates with IAP-predicted energy above hull >= 50 meV/atom
cd /crystal/HTS/7_EAH_prescreen
sh 0_setup.sh # download relevant entries for high-throughput energy above hull calculation
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.py
python 3_filter.pyRule out candidates with ALIGNN predicted band gap E_g < 0.1 eV (less likely to be a semiconductor)
cd /crystal/HTS/8_band_gap_prescreen
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.py!!! Note that VASP should be installed and POTCAR should be set up for pymatgen using "pmg config -p <EXTRACTED_VASP_POTCAR> <MY_PSP>" before performing this task. Because VASP is a commercial software, it is not installed in the docker image provided.
Perform geometry relaxation and band structure calculation at PBE level using VASP
cd /crystal/HTS/9_EAH_Band_gap_PBE
cp /crystal/HTS/7_EAH_prescreen/competitive_compositions.json.gz ./
python 1_splitRun.py # wait for jobs to finish (using qstat to check)
python 2_collect_clean_glob_details.py
python 3_filter.py # check results_7_EAH_prescreenfiltered_0.05eV.csv for details of promising candidates; check ./candidates for band structurescd invcryrep
python setup.py install@article{xiao2023invertible, title={An invertible, invariant crystallographic representation for inverse design of solid-state materials using generative deep learning}, author={Xiao, Hang and Li, Rong and Shi, Xiaoyang and Chen, Yan and Zhu, Liangliang and Wang, Lei and Chen, Xi}, year={2023} }