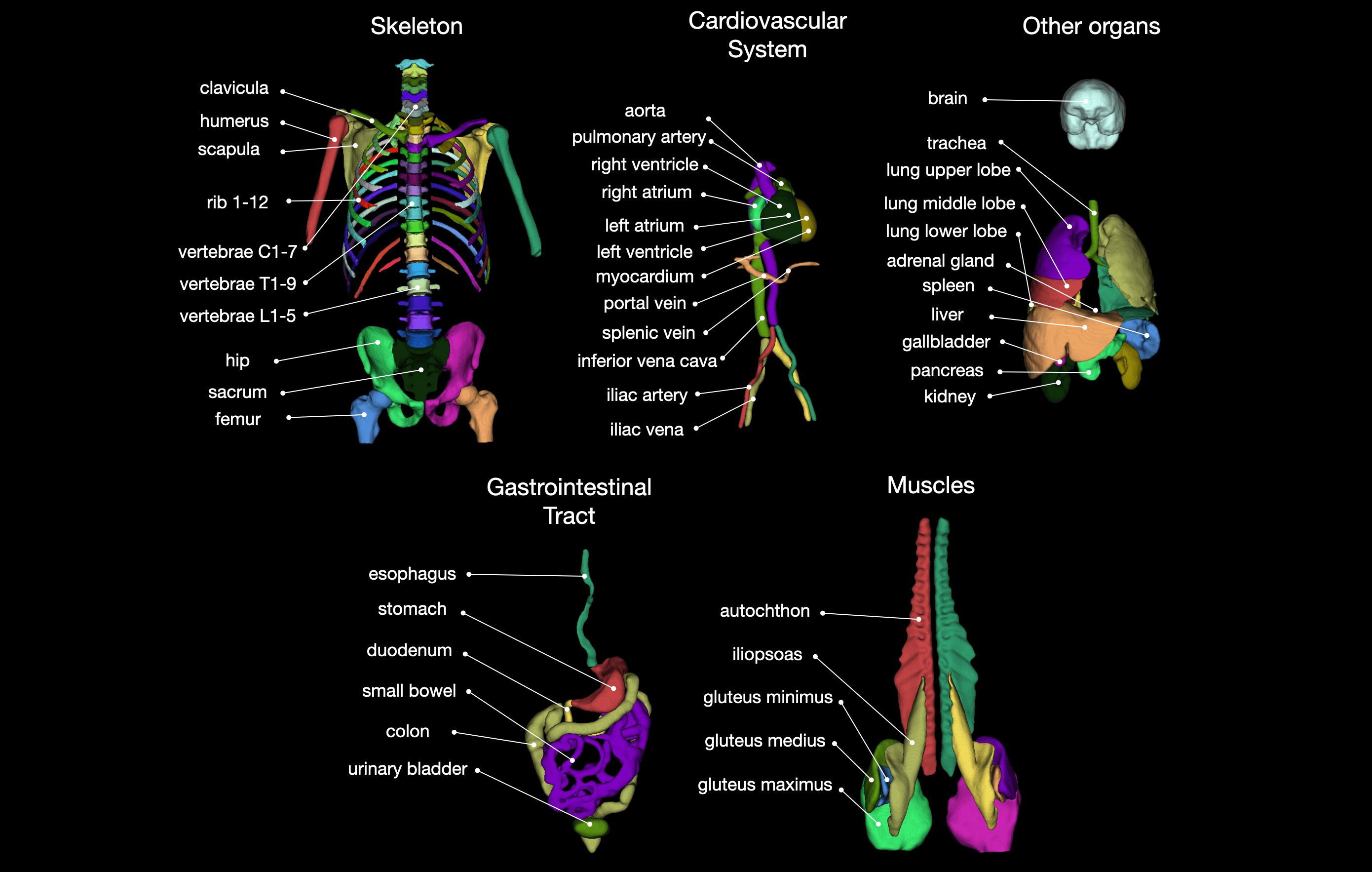
Tool for segmentation of 104 classes in CT images. It was trained on a wide range of different CT images (different scanners, institutions, protocols,...) and therefore should work well on most images. The training dataset with 1204 subjects can be downloaded from Zenodo. You can also try the tool online at totalsegmentator.com.
If you use it please cite our paper: https://arxiv.org/abs/2208.05868.
Install dependencies:
- Pytorch
- You should not have any nnU-Net installation in your python environment since TotalSegmentator will install its own custom installation.
Install Totalsegmentator
pip install TotalSegmentator
TotalSegmentator -i ct.nii.gz -o segmentations --fast --preview
Note: TotalSegmentator only works with a NVidia GPU. If you do not have one you can try our online tool: www.totalsegmentator.com
--fast: For faster runtime and less memory requirements use this option. It will run a lower resolution model (3mm instead of 1.5mm).--preview: This will generate a 3D rendering of all classes, giving you a quick overview if the segmentation worked and where it failed (seepreview.pngin output directory).--statistics: This will generate a filestatistics.jsonwith volume and mean intensity of each class.--radiomics: This will generate a filestatistics_radiomics.jsonwith radiomics features of each class. You have to install pyradiomics to use this (pip install pyradiomics).
We also provide a docker container which can be used the following way
docker run --gpus 'device=0' --ipc=host -v /absolute/path/to/my/data/directory:/workspace totalsegmentator:master TotalSegmentator -i /workspace/ct.nii.gz -o /workspace/segmentations
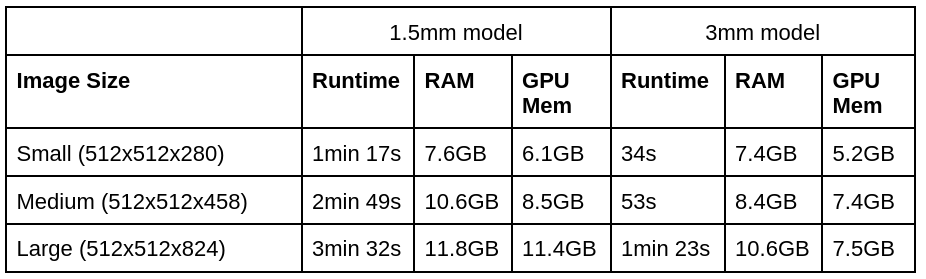
Totalsegmentator has the following runtime and memory requirements:
(1.5mm is the normal model and 3mm is the --fast model)
The exact split of the dataset can be found in the file meta.csv inside of the dataset. This was used for the validation in our paper.
The exact numbers of the results for the high resolution model (1.5mm) can be found here. The paper shown these numbers in the supplementary materials figure 11.
If you want to combine some subclasses (e.g. lung lobes) into one binary mask (e.g. entire lung) you can use the following command:
totalseg_combine_masks -i totalsegmentator_output_dir -o combined_mask.nii.gz -m lung
For more details see this paper https://arxiv.org/abs/2208.05868. If you use this tool please cite it as follows
Wasserthal J., Meyer M., Breit H., Cyriac J., Yang S., Segeroth M. TotalSegmentator: robust segmentation of 104 anatomical structures in CT images, 2022. URL: https://arxiv.org/abs/2208.05868. arXiv: 2208.05868
Moreover, we would really appreciate if you let us know what you are using this tool for. You can also tell us what classes we should add in future releases. You can do so here.