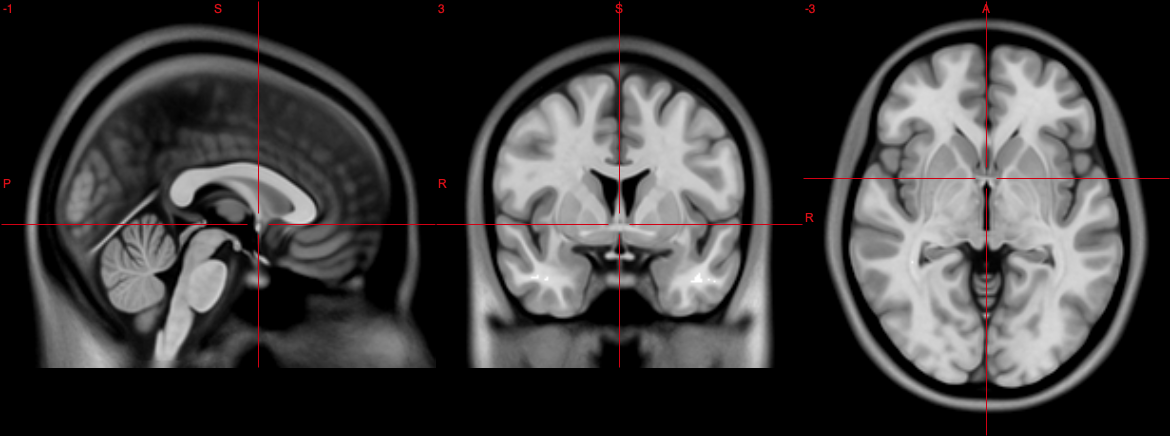
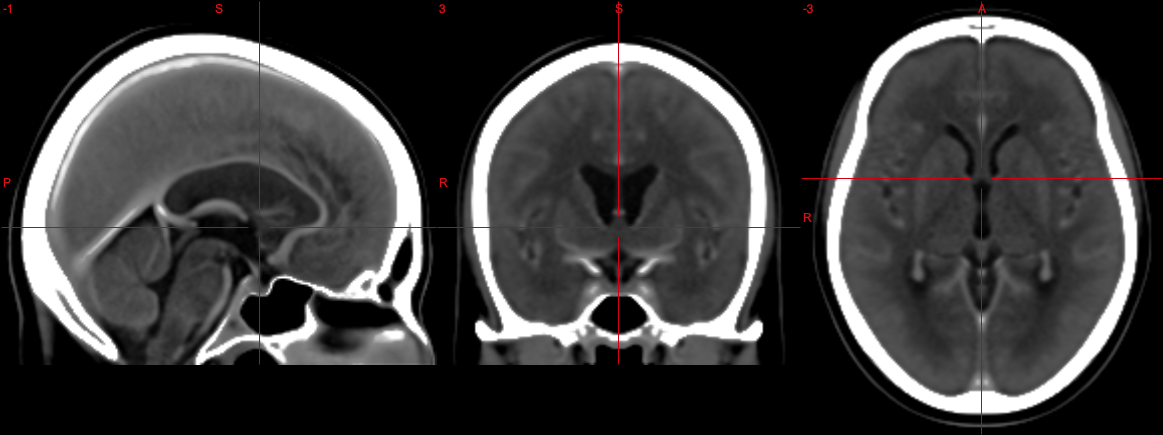
Everyday CT or MR examinations are supposed to be acquired in the same fashion, but slight deviations (e.g. due to subject/patient movement, differences in planning standards for MR and CT, or improper planning) may result in not easily comparable images. The fully automatic tilting of brainscans to Anterior Commissure - Posterior Commissure (fatbACPC) tool is meant to align CT and MR scans to the ACPC line to yield easily comparable images.
fatbACPC is meant to be used as a submodule of BrainSTEM.
Please note, this software is research-only and must not be used for clinical decisions/diagnosis.
The fatbACPC script takes a directory with DICOM files of a CT or (3D) MR brain scan as input and then aligns that scan with a reference volume, such as the MNI brain atlas.
The Anterior Commissure - Posterior Commissure (ACPC) line has been adopted as a standard in neuroimaging as a reference plane for axial imaging. Most atlases are aligned with the ACPC line and therefore this script should yield image volumes aligned to the ACPC line. Hence the name of the script.
The ACPC line is similar to the orbitomeatal line used as a common reference plane in CT, which is about 9° steeper than the ACPC line. Aligning both CT and MR scans to the ACPC line should yield image volumes, which are easily comparable longitudinally and between modalities.
Especially interesting for CT scans is the option to automatically generate mean slabs. By default, mean slabs of 5 mm thickness are generated for CT scans. When generating the slabs, an algorithm finds the top non-air slice in the volume and starts the creation of slabs with that slice.
A simple check for a mis-registration is in place. If rotation around one axis is more than 45°, we assume a mis-registration happened and try to realign the image volume after skull stripping. FSL's skull stripping might introduce it's own set of artifacts in CT, therefore, we apply it as "rescue attempt" only.
The aligned scans will be automatically exported back to the PACS.
Options to fatbACPC.bash:
-i --input [arg] Directory containing the DICOM input files. Required.
-k --keep-workdir After running, copy the temporary work directory into the input directory.
-c --cleanup After running, empty the source directory (reference DICOM and translation matrix are kept)
-p --no-pacs Do not send the results to the PACS.
-v Enable verbose mode, print script as it is executed.
-d --debug Enables debug mode.
-h --help This page.
We are listing the software versions we used (likely the latest available during development), but we are currently not aware that we rely on any features that are specific to one of the software's versions.
- BrainSTEM
- BASH, we used v4.4.19(1)
- bc, we used 1.07.1
- FSL, we used v5.0.11
- In the current early state: Git, we used v2.17.1
- parallel, we used v20180822
- Python 2 or 3, we used v2.7.15
fatbACPC is in the modules/ subdirectory of BrainSTEM (after you initialized and updated the submodules).
Copy the setup templates:
$ cd modules/fatbACPC
$ cp setup.fatbACPC.bash-template setup.fatbACPC.bashUpdate the PATH to FSL in setup.fatbACPC.bash:
# Setup FSL
#
export FSLDIR="/path/to/fsl-5.0.11"By default, mean slabs are only generated for CT examinations.
# Intended slice thickness for DICOM export in millimetres (mm),
# which will be matched as close as possible.
#
# Use -1 to not generate any mean slabs, for example on isometric
# MRI acquisitions.
#
intended_slice_thickness_ct=5
intended_slice_thickness_t1=-1
intended_slice_thickness_t2=-1Make sure that you enable fatbACPC in BrainSTEM.
Please see https://github.com/BrainImAccs/BrainSTEM#debugging
Please see templates/README.md for information on the templates.
The main scripts are based on the BASH3 Boilerplate.