Principal Component Anlaysis (PCA) in PyTorch. The intention is to provide a
simple and easy to use implementation of PCA in PyTorch, the most similar to
the sklearn's PCA as possible (in terms of API and, of course, output).
Plus, this implementation is fully differentiable and faster (thanks to GPU parallelization)!
Github repository: https://github.com/valentingol/torch_pca
Pypi project: https://pypi.org/project/torch_pca/
Documentation: https://torch-pca.readthedocs.io/en/latest/
Simply install it with pip:
pip install torch-pcaExactly like sklearn.decomposition.PCA but it uses PyTorch tensors as input and output!
from torch_pca import PCA
# Create like sklearn.decomposition.PCA, e.g.:
pca_model = PCA(n_components=None, svd_solver='full')
# Use like sklearn.decomposition.PCA, e.g.:
>>> new_train_data = pca_model.fit_transform(train_data)
>>> new_test_data = pca_model.transform(test_data)
>>> print(pca.explained_variance_ratio_)
[0.756, 0.142, 0.062, ...]More details and features in the API documentation.
Use the pytorch framework allows the automatic differentiation of the PCA!
The PCA transform method is always differentiable so it is always possible to compute gradient like that:
pca = PCA()
for ep in range(n_epochs):
optimizer.zero_grad()
out = neural_net(inputs)
with torch.no_grad():
pca.fit(out)
out = pca.transform(out)
loss = loss_fn(out, targets)
loss.backward()If you want to compute the gradient over the full PCA model (including the
fitted pca.n_components), you can do it by using the "full" SVD solver
and removing the part of the fit method that enforce the deterministic
output by passing determinist=False in fit or fit_transform method.
This part sort the components using the singular values and change their sign
accordingly so it is not differentiable by nature but may be not necessary if
you don't care about the determinism of the output:
pca = PCA(svd_solver="full")
for ep in range(n_epochs):
optimizer.zero_grad()
out = neural_net(inputs)
out = pca.fit_transform(out, determinist=False)
loss = loss_fn(out, targets)
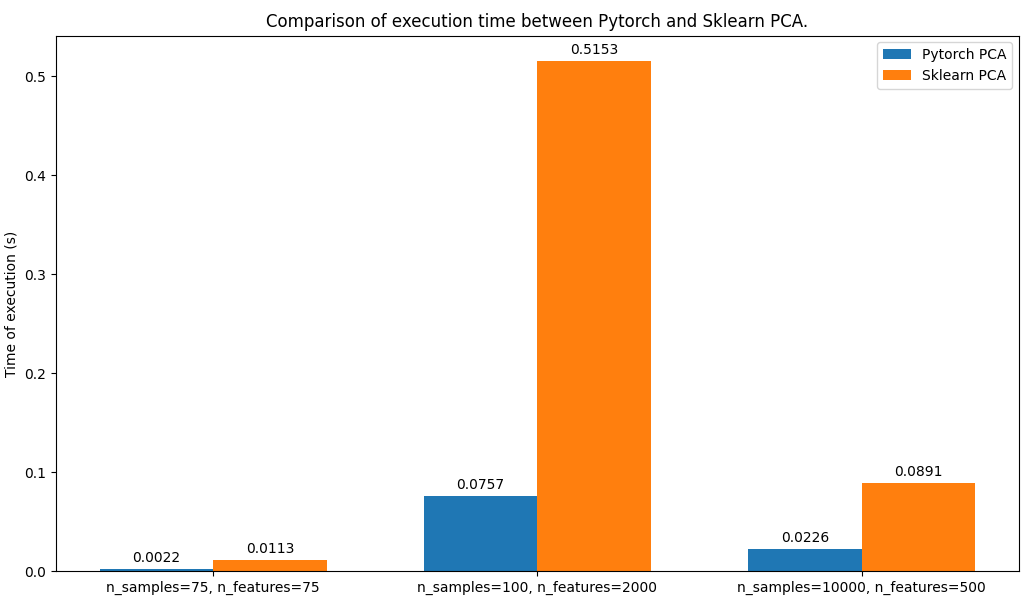
loss.backward()As we can see below the PyTorch PCA is faster than sklearn's PCA, in all the configs tested with the parameter by default (for each PCA model):
-
fit,transform,fit_transformmethods. - All attributes from sklean's PCA are available:
explained_variance_(ratio_),singular_values_,components_,mean_,noise_variance_, ... - Full SVD solver
- SVD by covariance matrix solver
- Randomized SVD solver
- (absent from sklearn) Decide how to center the input data in
transformmethod (default is like sklearn's PCA) - Find number of components with explained variance proportion
- Automatically find number of components with MLE
-
inverse_transformmethod - Whitening option
-
get_covariancemethod -
get_precisionmethod andscore/score_samplesmethods
- Support sparse matrices with ARPACK solver
Feel free to contribute to this project! Just fork it and make an issue or a pull request.
See the CONTRIBUTING.md file for more information.