 Code Author: Jonathan Crabbé (jc2133@cam.ac.uk)
Code Author: Jonathan Crabbé (jc2133@cam.ac.uk)
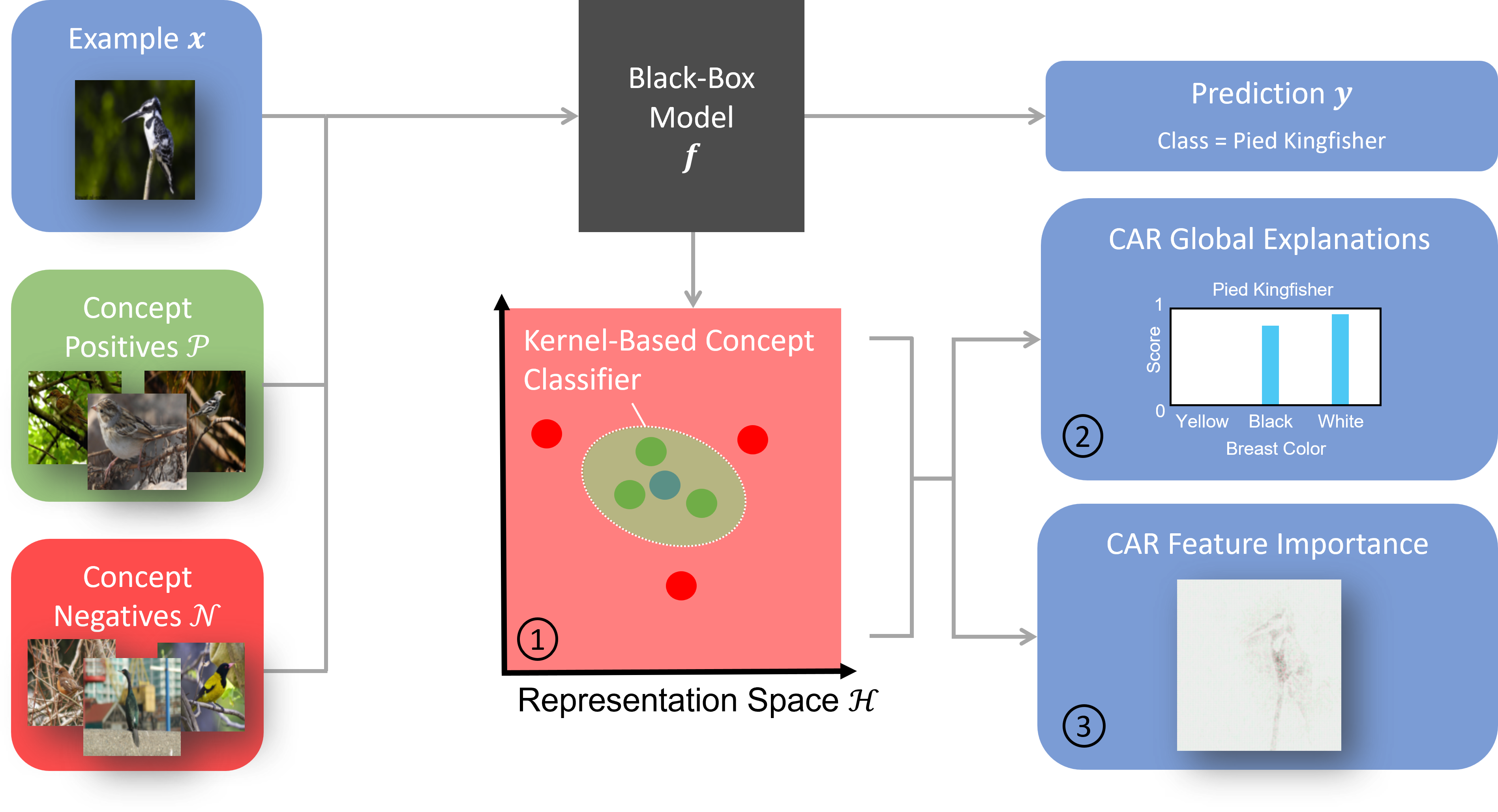
This repository contains the implementation of CARs, a framework to explain the latent representations of neural networks with the help of user-defined concepts. For more details, please read our NeurIPS 2022 paper: 'Concept Activation Regions: A Generalized Framework For Concept-Based Explanations'.
From repository:
- Clone the repository
- Create a new virtual environment with Python 3.9
- Run the following command from the repository folder:
pip install -r requirements.txtWhen the packages are installed, you are ready to perform concept explanations.
OR from to install from source, simply perform:
pip install .
Bellow, you can find a toy demonstration where we compute label-free feature and example importance with a MNIST autoencoder. The relevant code can be found in the folder explanations.
import torch
from torchvision.datasets import MNIST
from torchvision.transforms import transforms
from pathlib import Path
from cars.utils.dataset import generate_mnist_concept_dataset
from cars.models.mnist import ClassifierMnist
from cars.experiments.mnist import concept_to_class
from cars.explanations.concept import CAR
from cars.explanations.feature import CARFeatureImportance
device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
data_dir = Path.cwd()/"data/mnist"
# Load model
model = ClassifierMnist(latent_dim=42).to(device)
# Load concept sets
X_train, C_train = generate_mnist_concept_dataset(concept_to_class["Loop"],
data_dir,
train=True,
subset_size=200,
random_seed=42)
X_test, C_test = generate_mnist_concept_dataset(concept_to_class["Loop"],
data_dir,
train=False,
subset_size=100,
random_seed=42)
# Evaluate latent representation
X_train = torch.from_numpy(X_train).to(device)
X_test = torch.from_numpy(X_test).to(device)
H_train = model.input_to_representation(X_train)
H_test = model.input_to_representation(X_test)
# Fit CAR classifier
car = CAR(device)
car.fit(H_train.detach().cpu().numpy(), C_train)
car.tune_kernel_width(H_train.detach().cpu().numpy(), C_train)
# Predict concept labels
C_predict = car.predict(H_test.detach().cpu().numpy())
# Load test set
test_set = MNIST(data_dir, train=False, download=True)
test_set.transform = transforms.Compose([transforms.ToTensor()])
test_loader = torch.utils.data.DataLoader(test_set, batch_size=100, shuffle=False)
# Evaluate feature importance on the test set
baselines = torch.zeros([1, 1, 28, 28]) # Black image baseline
car_features = CARFeatureImportance("Integrated Gradient", car, model, device)
feature_importance = car_features.attribute(test_loader, baselines=baselines)Run the following script
python -m experiments.mnist --name experiment_name --train --plotwhere the --train option should only be used one time to fit a model for all the experiments. The experiment_name parameter can take the following values:
| experiment_name | description |
|---|---|
| concept_accuracy | Consistency check for CAR classifiers (paper Section 3.1.1) |
| statistical_significance | Statistical significance for the concepts (paper Section 3.1.1) |
| global_explanations | Consistency check for TCAR explanations (paper Section 3.1.2) |
| feature_importance | Consistency check for feature importance (paper Section 3.1.3) |
| kernel_sensitivity | Kernel sensitivity experiment (paper Appendix A) |
| concept_size_impact | Impact of concept set size (paper Appendix A) |
| tcar_inter_concepts | Intersection between concepts (paper Appendix B) |
| adversarial_robustness | Robustness w.r.t. adversarial perturbation (paper Appendix G) |
| senn | Experiment with self-explaining neural network (paper Appendix H) |
The resulting plots and data are saved here.
Our script automatically downloads the ECG dataset from Kaggle. To do so, one has to create a Kaggle token as explained here. Once the token is properly set-up, one can run our experiments with the script
python -m experiments.ecg --name experiment_name --train --plotwhere the --train option should only be used one time to fit a model for all the experiments. The experiment_name parameter can take the following values:
| experiment_name | description |
|---|---|
| concept_accuracy | Consistency check for CAR classifiers (paper Section 3.1.1) |
| statistical_significance | Statistical significance for the concepts (paper Section 3.1.1) |
| global_explanations | Consistency check for TCAR explanations (paper Section 3.1.2) |
| feature_importance | Consistency check for feature importance (paper Section 3.1.3) |
The resulting plots and data are saved here.
-
Create a folder data/cub in the repo folder.
-
Download the CUB dataset, the processed CUB dataset and the Place365 dataset.
-
Extract the two downloaded archives in the created folder data/cub
-
Process the CUB dataset for the robustness experiment with the script
python -m utils.robustnessOnce the dataset has been processed, one can run our experiments with the script
python -m experiments.cub --name experiment_name --train --plotwhere the --train option should only be used one time to fit a model for all the experiments. The experiment_name parameter can take the following values:
| experiment_name | description |
|---|---|
| concept_accuracy | Consistency check for CAR classifiers (paper Section 3.1.1) |
| statistical_significance | Statistical significance for the concepts (paper Section 3.1.1) |
| global_explanations | Consistency check for TCAR explanations (paper Section 3.1.2) |
| feature_importance | Consistency check for feature importance (paper Section 3.1.3) |
| tune_kernel | Kernel optimization experiment (paper Appendix A) |
| global_explanations_robustness | Robustness w.r.t. background shift (paper Appendix G) |
The resulting plots and data are saved here.
-
Create a folder data/seer in the repo folder.
-
Get the raw SEER Dataset in the form of a csv file
seer.csv. -
Put the file
seer.csvin the folder data/seer.
Once the dataset has been processed, one can run our experiments with the script
python -m experiments.seer --name experiment_name --train --plotwhere the --train option should only be used one time to fit a model for all the experiments. The experiment_name parameter can take the following values:
| experiment_name | description |
|---|---|
| concept_accuracy | Prostate cancer use case (paper Section 3.2) |
| global_explanations | Prostate cancer use case (paper Section 3.2) |
| feature_importance | Prostate cancer use case (paper Section 3.2) |
The resulting plots and data are saved here.
If you use this code, please cite the associated paper:
@misc{https://doi.org/10.48550/arxiv.2209.11222,
doi = {10.48550/ARXIV.2209.11222},
url = {https://arxiv.org/abs/2209.11222},
author = {Crabbé, Jonathan and van der Schaar, Mihaela},
keywords = {Machine Learning (cs.LG), Artificial Intelligence (cs.AI), FOS: Computer and information sciences, FOS: Computer and information sciences},
title = {Concept Activation Regions: A Generalized Framework For Concept-Based Explanations},
publisher = {arXiv},
year = {2022},
copyright = {Creative Commons Attribution 4.0 International}
}