PyTorch implementation of ICML'23 paper Learning Representations without Compositional Assumptions. Authors: Tennison Liu, Jeroen Berrevoets, Zhaozhi Qian, Mihaela van der Schaar
This work addresses the issue of unsupervised representation learning on tabular datasets that contain feature sets from various sources of measurements. Traditional methods, which tackle this problem using the multi-view framework, are constrained by predefined assumptions that assume feature sets share the same information and representations should learn globally shared factors. However, this assumption is not always valid for real-world tabular datasets with complex dependencies between feature sets, resulting in localized information that is harder to learn. To overcome this limitation, we propose a data-driven approach that learns feature set dependencies by representing feature sets as graph nodes and their relationships as learnable edges. We introduce
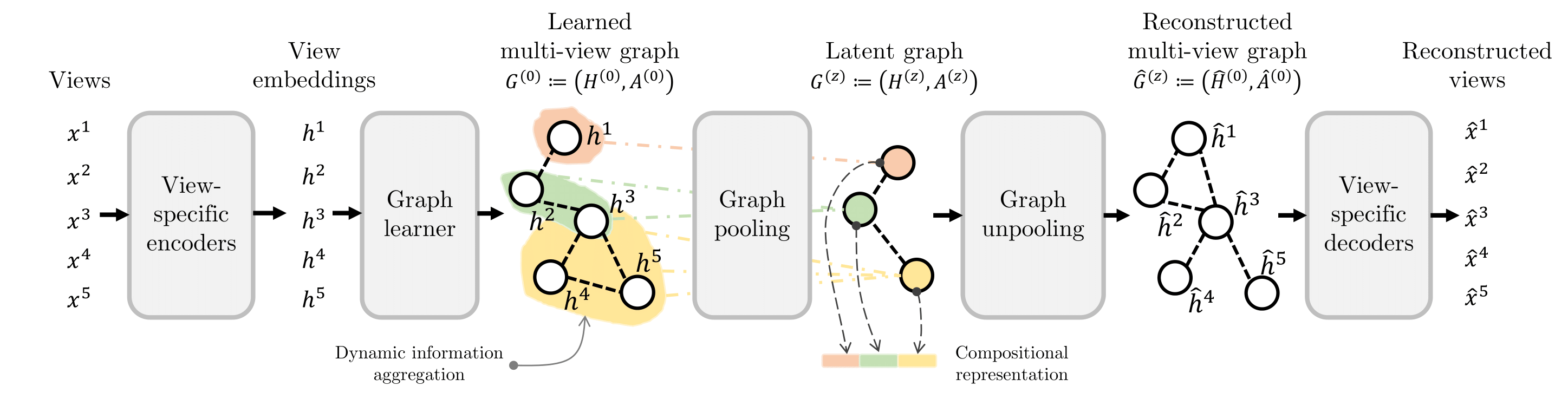 High-level illustration of LEGATO. The latent graph dynamically pools information by considering both view embeddings and dependencies. The latent graph returns a compositional representation for downstream tasks.
High-level illustration of LEGATO. The latent graph dynamically pools information by considering both view embeddings and dependencies. The latent graph returns a compositional representation for downstream tasks.
To setup the virtual environment and necessary packages, please run the following commands:
$ conda create --name legato_env python=3.8
$ conda activate legato_env
Clone this repository and navigate to the root directory:
$ git clone https://github.com/tennisonliu/LEGATO.git
$ cd LEGATO
Install the required modules:
$ pip install -r requirements.txt
Our algorithm is implemented in method/LEGATO.py, with hyperparameter optimization, training and evaluation scripts in training_utils/.
To reproduce results in the paper, see the corresponding scripts in exp/{exp_name}, e.g. exps/uci_exp. We have included experiment files for UCI Multiple Feature Sets (uci_exp), TCGA (tcga_exp), UK-Biobank (biobank_exp), and our simulation experiments (simulation_exp). Specifically, follow the following commands:
- Place dataset in
exps/datafolder. - Perform hyperparameter tuning using
training_utils/hyperopt.py. Save the best hyperparameters inexps/uci_exp/configs.json. Our experimental configurations are already loaded in the file. - Execute the training and evaluation scripts in
exps/uci_exp/run_uci_exp.py, which saves the results inexps/results.
To run experiments on your own datasets, modify any of the run_exp.py files based on your dataset and hyperparameters.
If our paper or code helped you in your own research, please cite our work as: