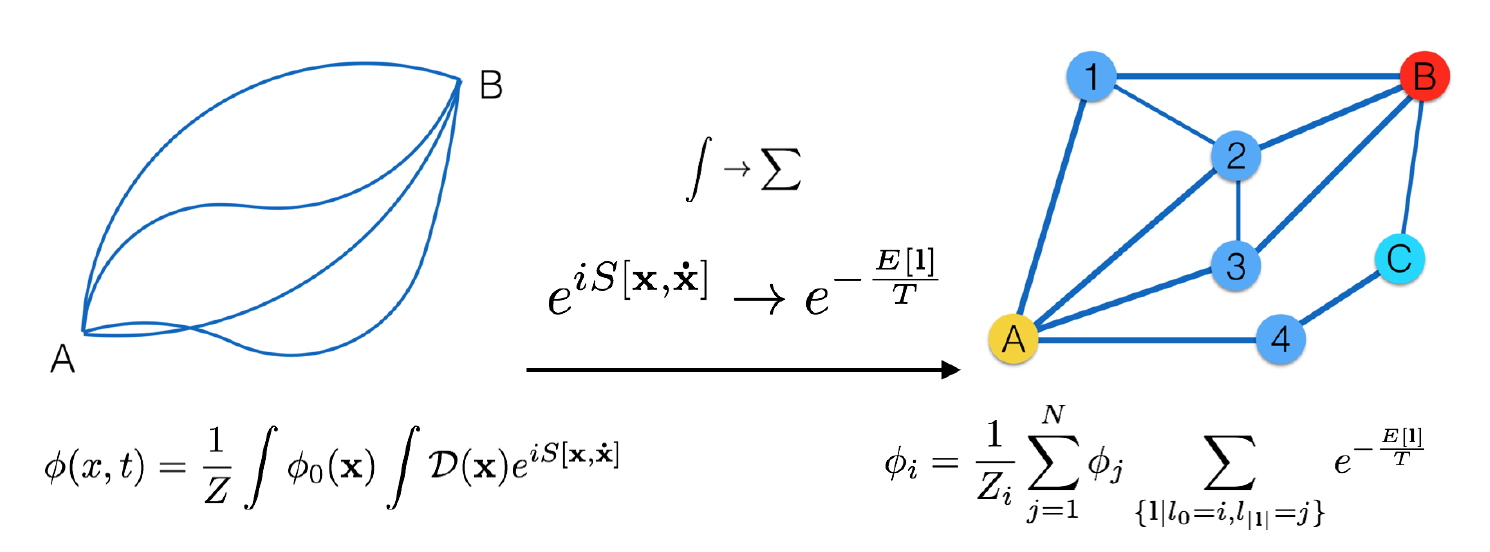This repository is the official implementation of Path Integral Based Convolution and Pooling for Graph Neural Networks.
In the figure above we draw the analogy between our discrete version and the original formulation. It is straightforward to see that the integral should now be replaced by a summation, and
To install requirements:
pip install -r requirements.txt
To train and test the model(s) in the paper, run the following command. We provide the codes for PAN on three benchmarks in Table 1 and the new dataset PointPattern. The dataset will be downloaded and preprocessed before training. All the experiments were performed using PyTorch Geometric and run on a server with Intel(R) Core(TM) i9-9820X CPU 3.30GHz, NVIDIA GeForce RTX 2080 Ti and NVIDIA TITAN V GV100.
PAN on Graph classification benchmark datasets; $dataname to be replaced by PROTEINS, PROTEINS_full, NCI1, AIDS, Mutagenicity
python pan_benchmark.py --dataset_name $dataname --L 3 --runs 10
PAN on PointPattern classification task
python pan_pointpattern.py --phi 0.3 --L 4 --runs 10
Other hyperparameters: --batch_size, --learning_rate, --weight_decay, --pool_ratio, --nhid, --epochs
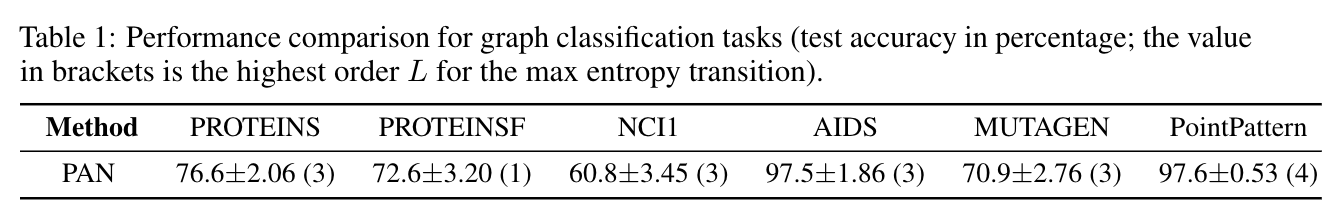
Our model PAN achieves the following performance on graph classification benchmark datasets MUTAG, PROTEINSF and NCI1, and our new graph classification dataset PointPattern (with phi=0.35). The table below shows the mean test accuracy with SD for 10 repetitions. Compared to existing methods such as GCNConv+TopKPool, SAGEConv+SAGPool, GATConv+EdgePool, with the same network architecture, the PAN achieves top test accuracy on most of these datasets. The results are obtained using the above .py programs. The test results on other benchmarks and comparison with the results of other methods can be seen in the paper.
Our new datasets are for 3-classification task of point distribtuion graphs, stemming from statistical mechanics. Use following commands to download the three typical datasets we used in the paper:
import os
import zipfile
import gdown
phi = 0.3 # change to 0.35 or 0.4 for other two PointPattern datasets
if phi==0.3:
ld_dir = 'hpr_phi03' + '_' + str(num_graph) + '/'
url = 'https://drive.google.com/uc?id=1C3ciJsteqsKFVGF8JI8-KnXhe4zY41Ss'
output = 'hpr_phi03' + '_' + str(num_graph) + '.zip'
if phi==0.4:
ld_dir = 'hpr_phi04' + '_' + str(num_graph) + '/'
url = 'https://drive.google.com/uc?id=1rsTh09FzGxHculBVrYyl5tOHD9mxqc0G'
output = 'hpr_phi04' + '_' + str(num_graph) + '.zip'
if phi==0.35:
ld_dir = 'hpr_phi035' + '_' + str(num_graph) + '/'
url = 'https://drive.google.com/uc?id=16pI974P8WzanBUPrMHIaGfeSLoksviBk'
output = 'hpr_phi035' + '_' + str(num_graph) + '.zip'
if not os.path.exists(output):
gdown.download(url, output, quiet=False)
with zipfile.ZipFile(output, 'r') as zip_ref:
zip_ref.extractall()
os.remove(output)
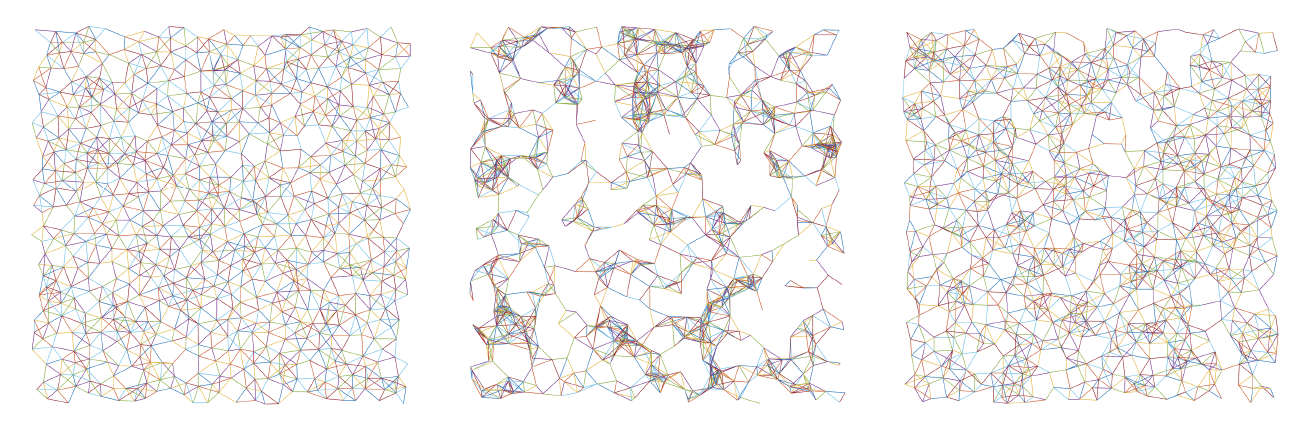
The following shows examples of graphs in PointPattern, phi=0.3; left to right: HS, Poisson, RSA
If you have used our codes and datasets, please cite:
@inproceedings{ma2020path,
title={Path Integral Based Convolution and Pooling for Graph Neural Networks},
author={Ma, Zheng and Xuan, Junyu and Wang, Yu Guang and Li, Ming and Lio, Pietro},
booktitle={NeurIPS},
year={2020}
}
Copyright (c) <2020>
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.