| Platforms | OS | R CMD check | Coverage |
|---|---|---|---|
| Travis CI | Linux | ||
| Bioc (devel) | Multiple | NA |
|
| Bioc (release) | Multiple | NA |
The iSEE package provides an interactive user interface for exploring data in objects derived from the SummarizedExperiment class.
Particular focus is given to single-cell data stored in the SingleCellExperiment derived class.
The user interface is implemented with RStudio's Shiny, with a multi-panel setup for ease of navigation.
This initiative was proposed at the European Bioconductor Meeting in Cambridge, 2017. Current contributors include:
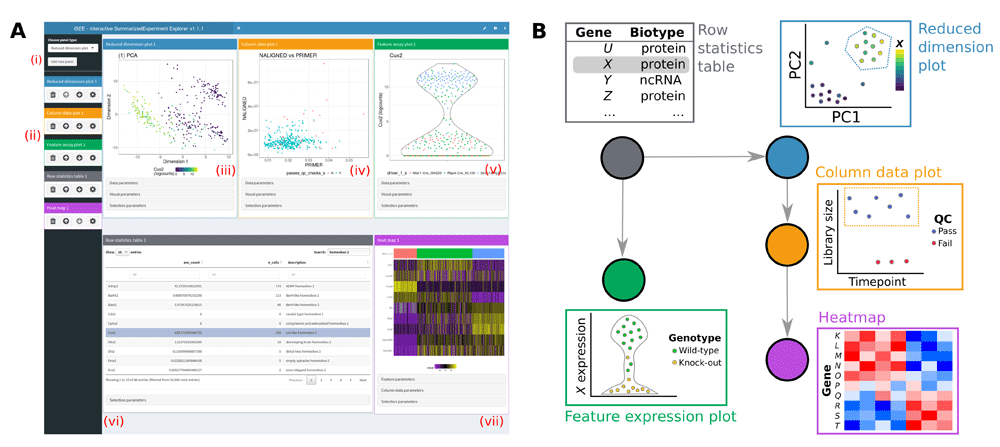
The user interface of iSEE web-applications currently offers the following features:
✅ Multiple interactive plot types with selectable points.
✅ Interactive tables with selectable rows.
✅ Coloring of samples and features by metadata or expression data.
✅ Zooming to a plot subregion.
✅ Transmission of point selections between panels to highlight, color, or restrict data points in the receiving panel(s).
✅ Lasso point selection to define complex shapes.
The iSEE user interface currently contains the following components where each data point represents a single biological sample:
✅ Reduced dimension plot: Scatter plot of reduced dimensionality data.
✅ Column data plot: Adaptive plot of any one or two sample metadata. A scatter, violin, or square design is dynamically applied according to the continuous or discrete nature of the metadata.
✅ Feature assay plot: Adaptive plot of expression data across samples for any two features or one feature against one sample metadata.
✅ Column statistics table: Table of sample metadata.
The iSEE user interface currently contains the following components where each data point represents a genomic feature:
✅ Row data plot: Adaptive plot of any two feature metadata. A scatter, violin, or square design is dynamically applied according to the continuous or discrete nature of the metadata.
✅ Sample assay plot: Adaptive plot of expression data across features for any two samples or one sample against one feature metadata.
✅ Row statistics table: Table of feature metadata.
The iSEE user interface contains the following components that integrate sample and feature information:
✅ Heat map plot: Visualize multiple features across multiple samples annotated with sample metadata.
The iSEE user interface allows users to programmatically define their own plotting and table panels.
✅ Custom data plot: Plotting panel that can be assigned any user-defined function returning a ggplot object.
✅ Custom statistics table: Table panel that can be assigned any user-defined function returning a data.frame object.
✅ The iSEE user interface continually tracks the code corresponding to all visible plotting panels. This code is rendered in a shinyAce text editor and can be copy-pasted into R scripts for customization and further use.
✅ Speech recognition can be enabled to control the user interface using voice commands.
We set up instances of iSEE applications running on diverse types of datasets at those addresses:
- http://shiny.imbei.uni-mainz.de:3838/iSEE
- https://marionilab.cruk.cam.ac.uk/iSEE_allen
- https://marionilab.cruk.cam.ac.uk/iSEE_tcga
- https://marionilab.cruk.cam.ac.uk/iSEE_pbmc4k
- https://marionilab.cruk.cam.ac.uk/iSEE_cytof
Please keep in mind that those public instances are for trial purposes only;
yet they demonstrate how you or your system administrator can setup iSEE for analyzing or sharing your precomputed SummarizedExperiment/SingleCellExperiment object.
If you want to extend the functionality of iSEE, you can create custom panels which add new possibilities to interact with your data. You can find a gallery with working examples of how to do it here. Feel free to contact the developing team, should you need some clarifications on how iSEE works internally. Submit a pull request once the implementation is complete, if you want to have it added to the gallery.