In this repository we implement a novel evolutionary algorithm, EComp, for compressing neural networks. It also includes testing facilities to investigate the resulting compressed networks and the values obtained during optimization.
Starting from some parent network you would like to compress, EComp uses an evolutionary strategy with a multiobjective fitness function to search for neural architectures that are smaller in size (i.e. number of nodes in hidden layers). At the heart of this fitness function is the similarity loss, a custom loss objective inspired by t-SNE that quantifies the quality of a compressed network's hidden representations as compared to that of the parent network.
We show that a multiobjective fitness function yields the best results in achieving a balance between test accuracy and compressed size when we employ the similarity loss, as compared to fitness functions that rely solely on accuracy and/or compression rate. For an in-depth description of our experimental findings, please see our report.
The main library is executable via the main.py file. It currently runs optimizations on the FashionMNIST dataset. The hyperparameters can be configured inside main.py using the Configs object. The default values as defined in main.py and used for our experiments are:
configs = Configs(
POP_SIZE=100,
MUTATION_RATE=0.01,
EMB_LAYERS=["layer_1_act", "layer_2_act"],
RECOMBINATION_LAYERS=["layer_1_linear", "layer_2_linear", "output"],
EPOCHS=1,
LOSS_WEIGHTS=[0,2,1]
)Explanations of these parameters can be found in config.py or in our report.
Before running this library, make sure you install the necessary requirements with
pip install -r requirements.txtPlease run everything from the root of this repository to ensure saving works correctly. Assuming you have configured the Configs in main.py or left them at their default values, you can now run experiments using the following scheme:
python src/main.py <n_runs> <experiment_name>n_runs denotes the number of runs you want to execute the experiment. Pass 1 to try it out, but to collect results you should use a higher number, like 10, such that randomly bad results can be averaged out. These tend to occur frequently due to the random initialisation of compressed networks, but we found that in 10 runs we usually only find 1 or 2 outliers.
All experiments are automatically saved. Therefore, provide the experiment_name of your choice when calling the script.
A full example of an experiment we conducted for our report thus looks like this:
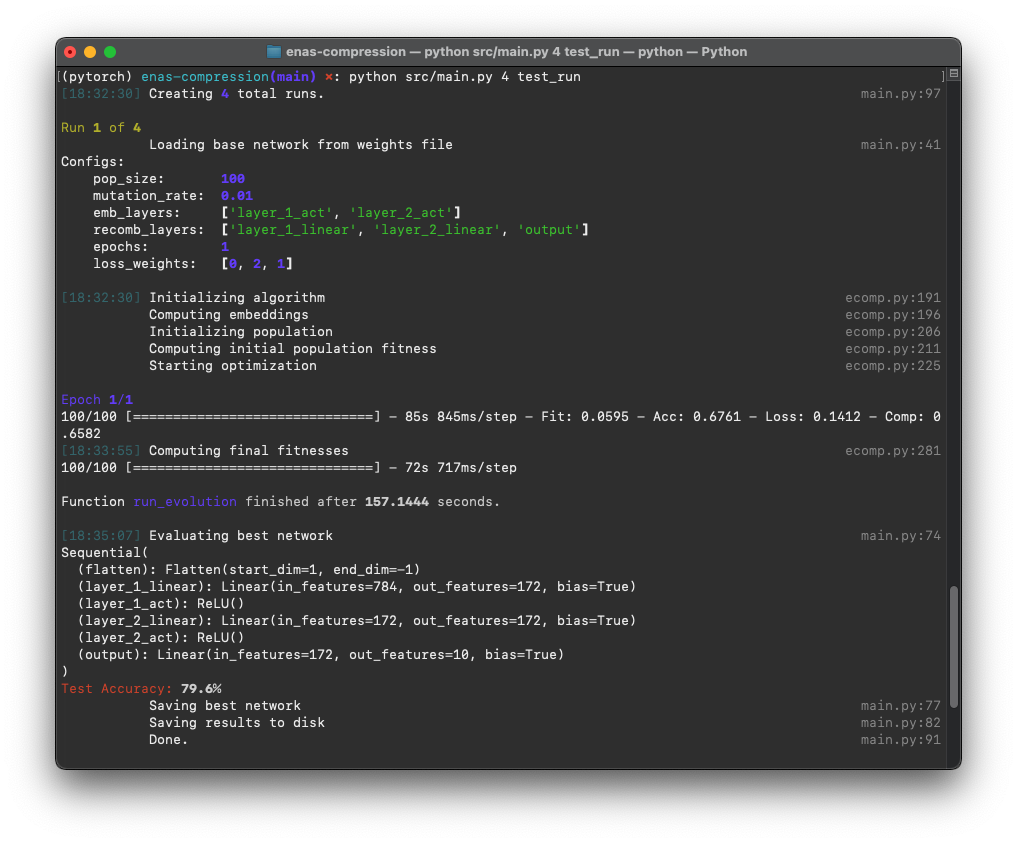
python src/main.py 10 weighting121Here is a preview of running it in a terminal:
The t-SNE inspired similarity loss is implemented as a class TSNELoss in src/losses.py. It needs to be instantiated with the embeddings of the origin network, which are the outputs obtained from a certain layer in the network for data fed into the network. Suppose you have 500 datapoints of irrelevant shape, and 256 neurons in the networks first hidden layer, the output of the first layer (the embeddings) should be of shape (500,256). Thus, this would be the shape required of the embeddings fed to the init method of TSNELoss.
Once the loss is instantiated, you can simply call it with the compressed network's embeddings, similarly obtained. The requirement here is that you have the same datapoints, as otherwise comparing embeddings does not make sense. It is however not required for the embeddings to have the same shape as the origin's in the second dimension, as only the amount of datapoints has to fit. Thus, to stick with our example from above, you may obtain embeddings from a compressed network with 128 neurons in its first hidden layer, which are consequently of shape (500,128).
Below is a rough example of how this would look in practice:
# Get the embeddings for a batch of data from the origin network
origin_embeddings = # output of hidden layer in origin network
# Instantiate the loss function with origin embeddings
loss_fn = TSNELoss(origin_embeddings)
# Get the embeddings for the same data from the compressed network
compressed_embeddings = # output of hidden layer in compressed network
# Get a single scalar loss value, corresponding to the informational
# similarity of the compressed networks hidden layer to that of the
# origin network
loss_value = loss_fn(compressed_embeddings)We note that EComp has only been tested as a proof-of-work on simple two-layer feed-forward neural networks using the FashionMNIST dataset. Although the results are very promising, we cannot predict how well they transfer to larger, more complex network architectures like CNNs, or more difficult datasets.
If you find EComp interesting and would like to use it or build on it, please let us now in the Issues section.
This project is licensed under the GNU General Public License v3.0.
- Thijme de Valk (tdevalk)
- Stijn van den Beemt (Stijnradboud)
- Pascal Schröder (verrannt)