This README.md was generated on 2024-05-23 by Yuting Vicky Lin (vicky.linyuting@gmail.com).
-
Author Information
-
First author
- Name: Dr. Yuting Vicky Lin
- Institution: Institute of Oceanography, National Taiwan University, Taipei 10617, Taiwan
-
Second author
- Name: Prof. Pierre-Alexandre Château
- Institution: Department of Marine Environment and Engineering, National Sun Yat-Sen University, Kaohsiung 80420, Taiwan
-
Third author
- Name: Prof. Yoko Nozawa
- Institution: Tropical Biosphere Research Center, University of the Ryukyus, Okinawa 905-0227, Japan
-
Fourth author
- Name: Prof. Chih-Lin Wei
- Institution: Institute of Oceanography, National Taiwan University, Taipei 10617, Taiwan
-
Fifth author
- Name: Dr. Rainer Ferdinand Wunderlich
- Institution: Department of Bioenvironmental Systems Engineering, National Taiwan University, Taipei 10617, Taiwan & 5INRAE, UR EABX, 33612 Cestas, France
-
Sixth & corresponding author
- Name: Prof. Vianney Denis
- Institution: Institute of Oceanography, National Taiwan University, Taipei 10617, Taiwan
- Email: vianneydenis@ntu.edu.tw
-
-
Date of data collection: 2016-2020
-
Geographic location of data collection: Taiwan, West Pacific
-
Funding sources that supported the collection of the data: Marine National Park of Taiwan, National Science and Technology Council of Taiwan, Ocean Affairs Council of Taiwan
-
Recommended citation for this dataset Lin YT, Château P-A, Nozawa Y, Wei C-L, Wunderlich RF, Denis V (2024), Data from: Drivers of coastal benthic communities in a complex environmental setting, Dryad, Dataset, https://doi.org/10.5061/dryad.j0zpc86nz
[Data and R script are also available through the GitHub repository https://github.com/NTU-FRELab/drivers-communities in order to replicate our analyses]
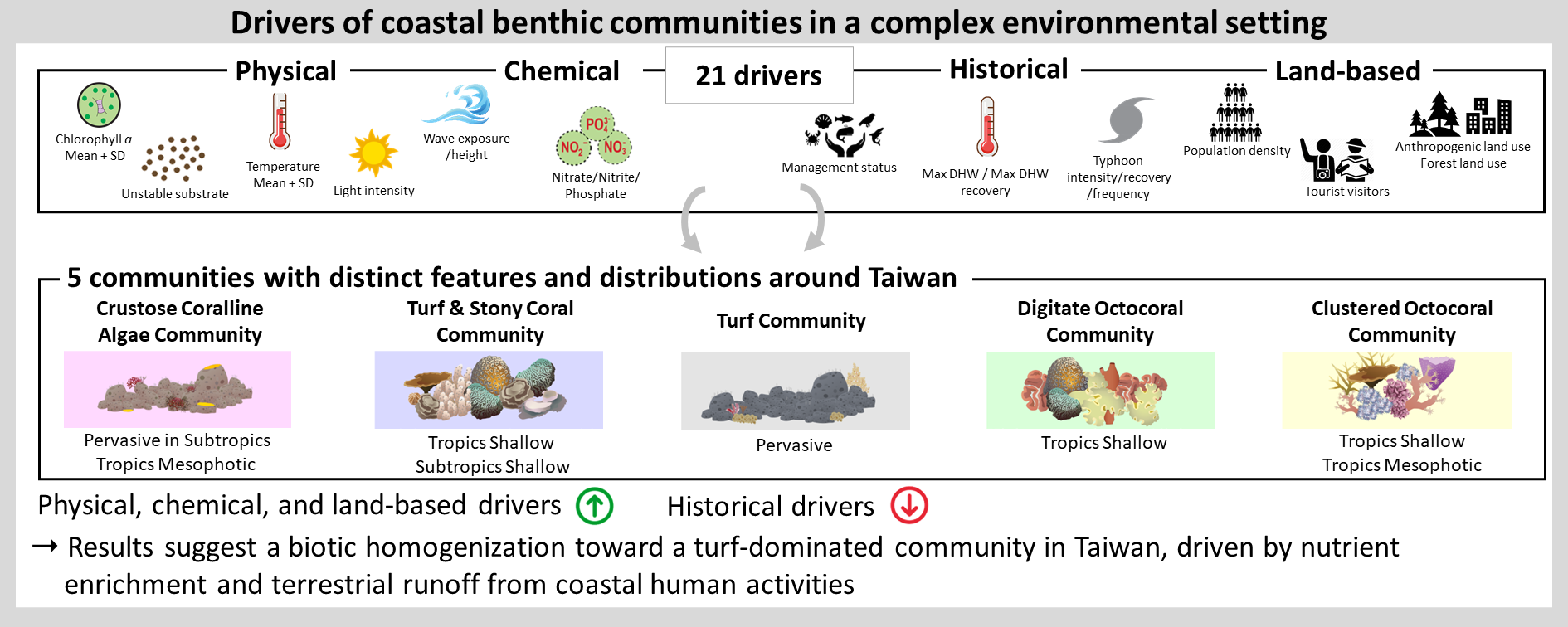
This research investigated how different environmental factors affect communities of marine organisms living on the seabed in coastal areas (i.e., benthic communities), which is important for deciding which factors need to be controlled. We collected data from 433 transects around Taiwan, including information on benthic composition and 21 environmental factors describing the physical environment, the chemicals in the water, the disturbance history and human activities on land. By analyzing these data, we were able to identify five types of communities dominated by different types of organisms, including crustose coralline algae, turf algae, stony corals, and digitate and bushy soft corals. We found that factors such as light intensity, nitrite levels, and population density were the most important in determining community types. Surprisingly, historical factors such as typhoons and abnormally high temperature had less influence. We also discovered that turf algae are widely distributed in Taiwan and are strongly promoted by human activities on land and nutrient enrichment. This suggests that human activities are making benthic communities in Taiwan more similar to each other, which could mask the effects of climate change and natural differences between regions. To protect these communities in Taiwan, it is important to control nutrient levels in the water and reduce pollution from the land. This will help these communities remain resilient in the face of climate change.
此研究探討了不同的環境因子如何影響生活在沿岸海床上的海洋生物群聚(即是底棲生物群聚)。這對於決定哪些因子需要被控制至關重要。此研究從台灣周圍的433條穿越線收集了數據,包括底棲生物群聚和21個環境因子(概括了物理環境、水中化學物質、歷史事件的干擾和陸地上的人類活動)的資料。通過分析這些數據,此研究確定了五種類型的群聚,這些群聚由不同類型的生物主導,包含殼狀珊瑚藻、絲狀藻類、石珊瑚、指狀和叢狀軟珊瑚。此研究發現,光照強度、亞硝酸鹽濃度和人口密度等因素在決定不同類型的群聚時相當重要。令人驚訝的是,颱風和溫度異常暖化等歷史事件的干擾影響並不大。此研究還發現,台灣周圍的絲狀藻類非常豐富,這主要是受到陸地上人類活動和水中營養鹽過高的強烈影響。這表示人類的行為正在使台灣周圍的群聚組成變得更加相似。這些人為活動的影響也可能掩蓋了底棲群聚對氣候變化的反應和地區間的自然差異。為了保護台灣的這些群聚,關鍵是要控制海水中的營養鹽、並減少陸地污染。這將有助於這些群聚在應對氣候變化時保持韌性。
-
Description of dataset
These data were used to identify distinct benthic communities around Taiwan and the important environmental drivers causing the difference.
-
File List
-
File 1: Linetal_dataset_Benthic.csv
- File 1 description: benthic cover estimates of the 433 sampling transects
-
File 2: Linetal_dataset_Env.csv
- File 2 description: data of 21 environmental drivers in the 433 sampling transects
-
File 3: Linetal_dataset_Cor.csv
- File 3 description: coordination and benthic information used to generate Figure 1b
-
File 4: Linetal_dataset_TW.shp
- File 4 description: file of Taiwan map used to generate Figure 1a
-
File 5: Linetal_dataset_TW.cpg
- File 5 description: file of Taiwan map used to generate Figure 1a
-
File 6: Linetal_dataset_TW.dbf
- File 6 description: file of Taiwan map used to generate Figure 1a
-
File 7: Linetal_dataset_TW.prj
- File 7 description: file of Taiwan map used to generate Figure 1a
-
File 8: Linetal_dataset_TW.shx
- File 8 description: file of Taiwan map used to generate Figure 1a
-
A detailed description of data acquisition and processing can be found in the published manuscript in the Marine Pollution Bulletin (https://doi.org/10.1016/j.marpolbul.2024.116462).
-
Number of variables/columns: 28
-
Number of cases/rows: 434
-
Missing data codes
None
-
Variable List
- Column A - transect
- Column B - ac_erect_single
- Column C - ag_articulated_calcareous
- Column D - ag_corticated_foliose
- Column E – ag_corticated_macrophyte
- Column F – cca_crustose
- Column G – cy_filamentous
- Column H – hc_arborescent
- Column I – hc_bushy
- Column J – hc_column
- Column K – hc_encrusting
- Column L – hc_foliose
- Column M – hc_massive
- Column N – hc_table
- Column O – oc_arborescent
- Column P – oc_bushy
- Column Q – oc_cluster
- Column R – oc_digitate
- Column S – oc_encrusting
- Column T – oc_lobate
- Column U – oc_massive
- Column V – sg
- Column W – sp_encrusting
- Column X – sp_massive
- Column Y – sp_repent
- Column Z – turf_filamentous
- Column AA – zo_encrusting
- Column AB – zo_massive
-
Abbreviations used
- for major benthic categories:
- ac: ascidian
- ag: alga
- cca: crustose coralline alga
- cy: cyanobacteria
- hc: hard coral
- oc: octocoral
- sg: sea grass
- sp: sponge
- turf:turf alga
- zo: zoanthid
- for major benthic categories:
-
Number of variables/columns: 26
-
Number of cases/rows: 434
-
Missing data codes
None
-
Variable List
- Column A - Region
- Column B - Site
- Column C - Depth
- Column D - Latitude
- Column E – Longitude
- Column F – Mean_SST
- Column G – SD_SST
- Column H – Light_intensity
- Column I – Wave_exposure
- Column J – Wave_height
- Column K – Mean_chl_a
- Column L – SD_chl_a
- Column M – Nitrate
- Column N – Nitrite
- Column O – Phosphate
- Column P – DHW
- Column Q – DHW_recovery
- Column R – Typhoon_disturbance
- Column S – Typhoon_recovery
- Column T – Typhoon_frequency
- Column U – Anthropogenic_land_use
- Column V – Forest_land_use
- Column W – Population_density
- Column X – Tourist_visitors
- Column Y – Unstable_substrate_cover
- Column Z – Management_status
-
Abbreviations used
- for environmental factors:
- Mean_SST: Mean sea surface temperature
- SD_SST: Standard deviation of sea surface temperature
- Light_intensity: Light intensity
- Wave_exposure: Wave exposure
- Wave_height: Wave height
- Mean_chl_a: Mean chlorophyll a
- SD_chl_a: Standard deviation of chlorophyll a
- DHW: Degree heating week
- DHW_recovery: Recovery time from the last extreme heat event
- Typhoon frequency: Number of severe typhoons in the last decade
- Typhoon disturbance: Maximum wind speed for the last severe typhoon
- Typhoon recovery: Recovery time from the last severe typhoon
- Anthropogenic_land_use: Anthropogenic land use
- Forest_land_use: Forest land use
- Population_density: Population density
- Tourist_visitors: Tourist visits
- Unstable_substrate_cover: Unstable substrate cover
- Management_status: Management status
- for environmental factors:
-
Number of variables/columns: 14
-
Number of cases/rows: 88
-
Missing data codes
None
-
Variable List
- Column A - Region
- Column B - Site
- Column C - Depth
- Column D - Latitude
- Column E – Longitude
- Column F – 1
- Column G – 2
- Column H – 3
- Column I – 4
- Column J – 5
- Column K – y
- Column L – x
- Column M – Label1
- Column N – Label2
-
Abbreviations used
- for variables:
- 1: CCA Community
- 2: Turf & Stony Coral Community
- 3: Turf Community
- 4: Digitate Community
- 5: Bushy Community
- y: the vertical position of the pie chart
- x: the horizontal position of the pie chart
- Label1: the number of sampling locations used in Figure 1a
- Label2: the number of sampling locations used in Figure 1b
- for variables:
R is required to open Lin_et_al_MPB.Rmd; the script was created using version 4.1.2. After opening the file, you can knit it to generate an HTML file and extract the R code. When knitting Lin_et_al_MPB.Rmd, make sure you have the following files available: Linetal_dataset_Benthic.csv, Linetal_dataset_Env.csv, Linetal_dataset_Cor.csv, Linetal_dataset_TW.shp, Linetal_dataset_TW.cpg, Linetal_dataset_TW.dbf, Linetal_dataset_TW.prj, and Linetal_dataset_TW.shx. Linetal_dataset_TW.cpg, Linetal_dataset_TW.dbf, Linetal_dataset_TW.prj, and Linetal_dataset_TW.shx are the companion files for Linetal_dataset_TW.shp.
Microsoft Excel can be used to view Linetal_dataset_Benthic.csv, Linetal_dataset_Env.csv, and Linetal_dataset_Cor.csv.