This repository provides instructions for downloading and preprocessing the EEGMMIDB dataset, a dataset commonly used for EEG (Electroencephalogram) analysis. The dataset is hosted on PhysioNet and contains EEG recordings from subjects performing motor imagery and executed movements.
To download the dataset, use the following command in the project root:
wget -r -N -c -np https://physionet.org/files/eegmmidb/1.0.0/This command uses wget to recursively download the necessary files from the PhysioNet repository.
Create a virtual environment and install the required packages by running the following commands:
python -m venv venv
source venv/bin/activate # On Windows, use `venv\Scripts\activate`
pip install -r requirements.txtThis ensures that you have a clean environment with all the dependencies.
Run the preprocessing script to generate preprocessed TensorFlow datasets:
python preprocessing.pyCreate the tf dataset running
python preprocessing.py
It is possible to change the values passed to each variable, but any changes will have to be adjusted in the model loader in the master.ipynb. Example:
python preprocessing.py --batch_size 2 --stride 100 --n_steps 700
This script handles the necessary steps to preprocess the EEGMMIDB dataset and prepares it for further analysis.
The getXYdata function extracts EEG signals (X) and labels (y) from the raw EEG recordings. The labels are encoded using LabelEncoder to convert categorical labels into a numerical format.
The splitTimeSeries function segments the time series data into instances with a specified number of steps (n_steps) and a specified stride. Choosing appropriate values for these parameters is crucial for subsequent analysis.
The generator_function iterates over EEG recordings of different individuals and exams using MNE to read raw EEG data. It calls getXYdata and splitTimeSeries to yield segmented instances for training.
The create_tf_dataset function converts the generator function into a TensorFlow dataset, specifying the output signature for X and y.
Command-line arguments such as --batch_size, --stride, and --n_steps impact the preprocessing pipeline. The optional --save argument allows for saving the full train and test datasets.
The full dataset is split into training and testing sets with an 80-20 split. Optionally, users can save the datasets using tf.data.experimental.save.
The --tiny argument allows for creating smaller datasets (tiny_train_dataset and tiny_test_dataset) useful for faster testing during development.
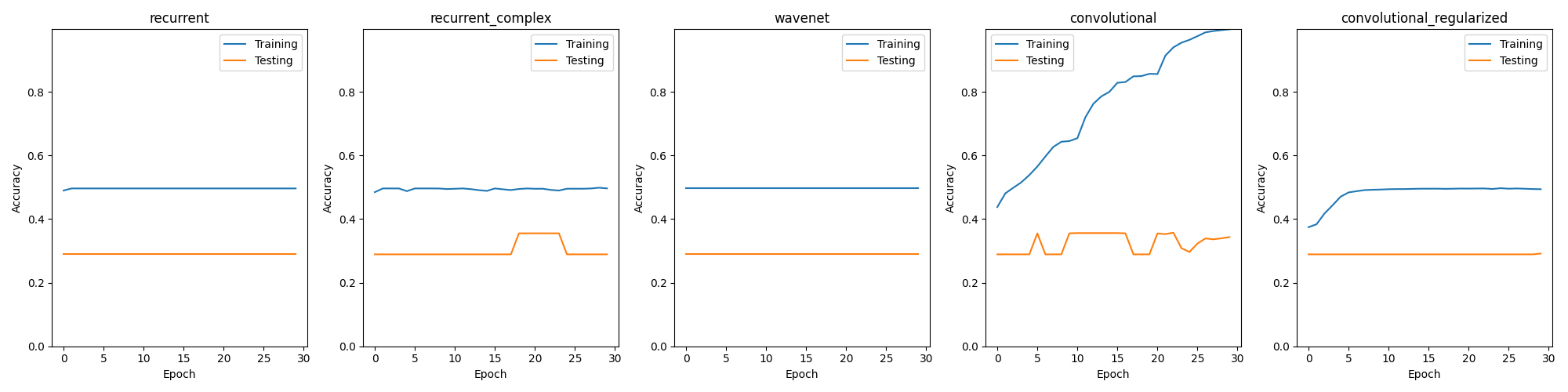
The following models were trained and tested with 5% of the original dataset separated into batches of 512 instances each, with a stride of 100 between the data points and 200 time steps per data point. Each model was trained with 30 epochs and the training and testing (validation) data do not overlap.
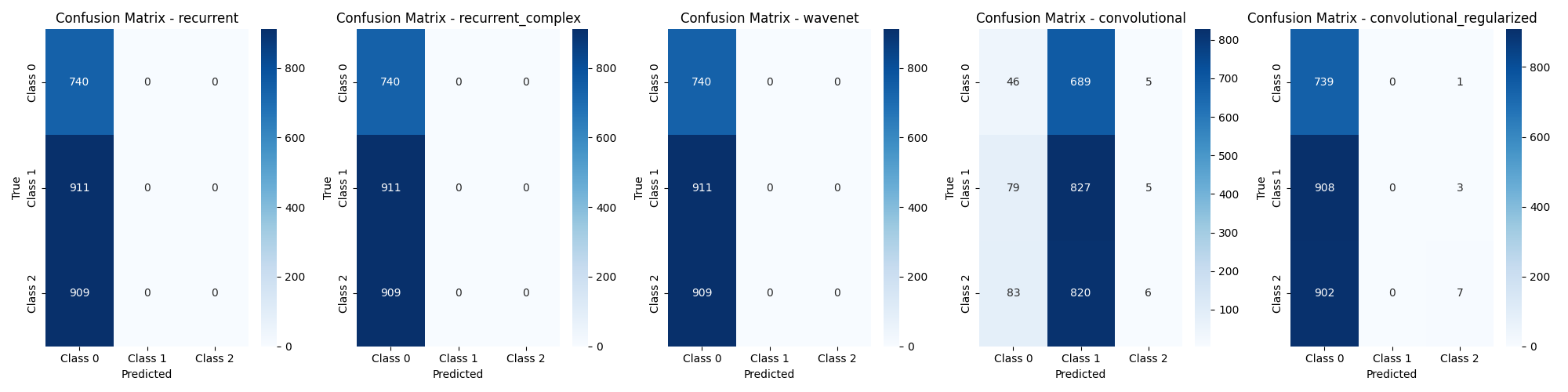
- Recurrent: a simple recurrent neural network was the first tested model, it clearly did not yield good results as it did underfitted severely and only predicted a single class on the test set.
- Recurrent Complex: a more complex recurrent neural network. It did also underfitted.
- Wavenet: Similar result to both recurrent models.
- Convolutional: A medium sized convolutional neural network. It overfitted and was the only model to predict different classes regularly over the dataset, all the other models only predicted a single class.
- Convolutional Regularized: In an attempt to stop the overfitting and improve performance over the test set the standard convolutional model was regulzarized using dropout layers, but it resulted in underfitting.
None of the models were able to predict the target variable with significant accuracy on the test set. The one which got the closest was the deep convolutional model.
The next steps will be:
- improving pre-processing: Adding fast fourier transform to the pre-processing layer and generating images from the dataset will greatly add to the depth of information and should improve drastically the performance of the convolutional models.
- Variational autoencoders: The state of the art papers in the field all use convolutional variational autoencoders to analyze and classify the resulting spectrograms from the fft, so these models might be the ideal tools to reach the envisioned goal of classifying motor imagery electro encephalogram datasets.
The initial models used in this project are sourced from the following repository:
https://github.com/ageron/handson-ml3/blob/main/15_processing_sequences_using_rnns_and_cnns.ipynb
These models serve as a starting point for further development and experimentation with the EEGMMIDB dataset.