Implementaion for "Adversarial Robust Training of Deep Learning MRI Reconstruction Models".
Code adapted from Official FastMRI and I-RIM for FastMRI.
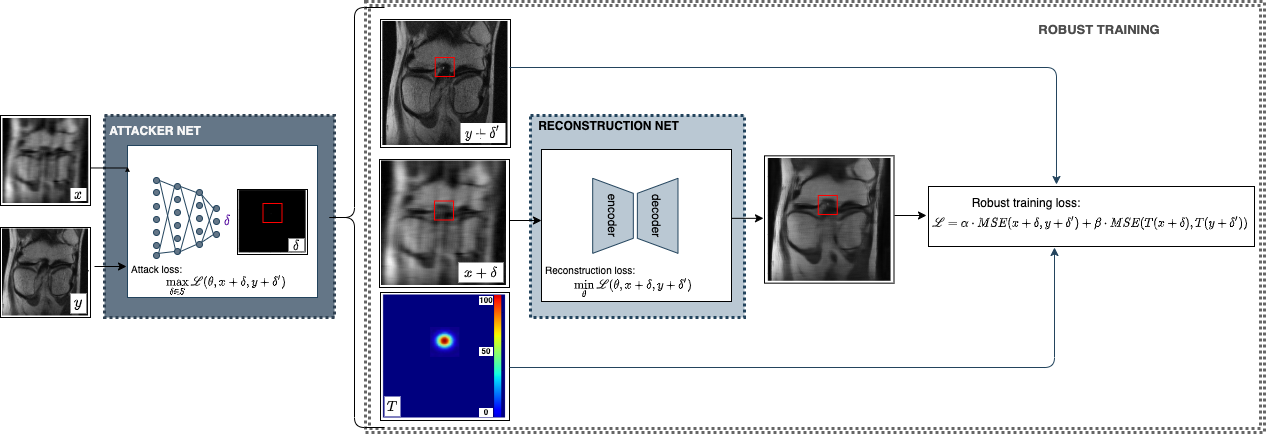
Deep Learning has shown potential in accelerating Magnetic Resonance Image acquisition and reconstruction. Nevertheless, there is a dearth of tailored methods to guarantee that the reconstruction of small features is achieved with high fidelity. In this work, we employ adversarial attacks to generate small synthetic perturbations that when added to the input MRI, they are not reconstructed by a trained DL reconstruction network. Then, we use robust training to increase the network's sensitivity to small features and encourage their reconstruction. Next, we investigate the generalization of said approach to real world features. For this, a musculoskeletal radiologist annotated a set of cartilage and meniscal lesions from the knee Fast-MRI dataset, and a classification network was devised to assess the features reconstruction. Experimental results show that by introducing robust training to a reconstruction network, the rate (4.8%) of false negative features in image reconstruction can be reduced. The results are encouraging and highlight the necessity for attention on this problem by the image reconstruction community, as a milestone for the introduction of DL reconstruction in clinical practice. To support further research, we make our annotation and implementation publicly available.
- See folders
unetandirim
Please download the "Fast-MRI dataset" using the official website. An example of MRI volume from the dataset is added in dataset
Once downloaded the dataset, please maintain its organization in two folders namely: singlecoil_train and singlecoil_val.
Download bounding box annotation.
cd unet
FNAF training:
python train_unet.py --gpu 3 --challenge singlecoil --data-path $DATA_PATH --exp-dir $EXPERIMENT_DIR --mask-type random --num-epochs 1000 --fn-train
BBox training:
python train_unet.py --gpu 3 --challenge singlecoil --data-path $DATA_PATH --exp-dir $EXPERIMENT_DIR --mask-type random --num-epochs 1000 --bbox_root $BBOX_ROOT_DIR
To start from pre-trained weights add to above:
--resume --checkpoint $CHECKPOINT
To evaluate FNAF attack on any U-Net:
4x:
python models/unet/train_unet.py --challenge singlecoil --mask-type random --accelerations 4 --center-fractions 0.08 --batch-size $BATCH_SIZE --fn-train --gpu $GPU_ID --data-path $DATA_PATH --exp-dir $EXPERIMENT_DIR --fnaf_eval $FNAF_ATTACK_RESULT_DIR --resume --checkpoint $CHECKPOINT
8x:
python models/unet/train_unet.py --challenge singlecoil --mask-type random --accelerations 8 --center-fractions 0.04 --batch-size $BATCH_SIZE --fn-train --gpu $GPU_ID --data-path $DATA_PATH --exp-dir $EXPERIMENT_DIR --fnaf_eval $FNAF_ATTACK_RESULT_DIR --resume --checkpoint $CHECKPOINT
Get non-attack reconstructions from U-Net:
See unet folder
cd irim
FNAF training:
python -m scripts.train_model \
--challenge singlecoil --batch_size 8 --n_steps 8 \
--n_hidden 64 64 64 64 64 64 64 64 64 64 64 64 \
--n_network_hidden 64 64 128 128 256 1024 1024 256 128 128 64 64 \
--dilations 1 1 2 2 4 8 8 4 2 2 1 1 \
--multiplicity 4 --parametric_output \
--loss ssim --resolution 320 --train_resolution 368 368 --lr_gamma 0.1 \
--lr 0.0001 --lr_step_size 30 --num_epochs 1000 --optimizer Adam \
--num_workers 4 --report_interval 100 --data_parallel
--data-path $DATA_PATH --exp_dir $EXPERIMENT_DIR --fnaf_train
BBox training:
python -m scripts.train_model \
--challenge singlecoil --batch_size 8 --n_steps 8 \
--n_hidden 64 64 64 64 64 64 64 64 64 64 64 64 \
--n_network_hidden 64 64 128 128 256 1024 1024 256 128 128 64 64 \
--dilations 1 1 2 2 4 8 8 4 2 2 1 1 \
--multiplicity 4 --parametric_output \
--loss ssim --resolution 320 --train_resolution 368 368 --lr_gamma 0.1 \
--lr 0.0001 --lr_step_size 30 --num_epochs 1000 --optimizer Adam \
--num_workers 4 --report_interval 100 --data_parallel
--data-path $DATA_PATH --exp_dir $EXPERIMENT_DIR --bbox_root $BBOX_ROOT_DIR
To start from pre-trained weights add to above:
--resume --checkpoint $CHECKPOINT
To evaluate FNAF attack on any I-RIM:
4x:
python -m scripts.train_model \
--challenge singlecoil --batch_size 24 --n_steps 8 \
--n_hidden 64 64 64 64 64 64 64 64 64 64 64 64 \
--n_network_hidden 64 64 128 128 256 1024 1024 256 128 128 64 64 \
--dilations 1 1 2 2 4 8 8 4 2 2 1 1 \
--multiplicity 4 --parametric_output \
--loss ssim --resolution 320 --train_resolution 368 368 --lr_gamma 0.1 \
--lr 0.0001 --lr_step_size 30 --num_epochs 1000 --optimizer Adam \
--num_workers 4 --report_interval 100 --data_parallel \
--data-path $DATA_PATH --resume --checkpoint $CHECKPOINT --val_accelerations 4 --val_center_fractions 0.08 --fnaf_eval $FNAF_ATTACK_RESULT_DIR
8x:
python -m scripts.train_model \
--challenge singlecoil --batch_size 24 --n_steps 8 \
--n_hidden 64 64 64 64 64 64 64 64 64 64 64 64 \
--n_network_hidden 64 64 128 128 256 1024 1024 256 128 128 64 64 \
--dilations 1 1 2 2 4 8 8 4 2 2 1 1 \
--multiplicity 4 --parametric_output \
--loss ssim --resolution 320 --train_resolution 368 368 --lr_gamma 0.1 \
--lr 0.0001 --lr_step_size 30 --num_epochs 1000 --optimizer Adam \
--num_workers 4 --report_interval 100 --data_parallel \
--data-path $DATA_PATH --resume --checkpoint $CHECKPOINT --val_accelerations 8 --val_center_fractions 0.04 --fnaf_eval $FNAF-ATTACK_RESULT_DIR
Get non-attack reconstructions from U-Net:
See irim folder
For U-Net and IRIM after getting the non-attack reconstructions (h5 files) :
python unet/common/evaluate.py --challenge singlecoil --target-path $TARGET_RECONSTRUCTION_PATH --predictions-path $PREDICTION_RECONSTRUCTION_PATH --bbox-root $BBOX_ROOT_DIR --by-slice
If you use this annotation for your research, please consider citing our paper.
@article{caliva2020adversarial,
title={Adversarial Robust Training in MRI Reconstruction},
author={Caliv{\'a}, Francesco and Cheng, Kaiyang and Shah, Rutwik and Pedoia, Valentina},
journal={arXiv preprint arXiv:2011.00070},
year={2020}
}