Quang Vinh Nguyen, Van Thong Huynh, Soo Hyung Kim
Official Tensorflow implementation
- (August 25, 2023)
- Paper accepted as ORAL at BMVC 2023 (Rank A) ! ⭐
- (May 12, 2023)
- Paper submitted at BMVC 2023 (Rank A) ! ⌚
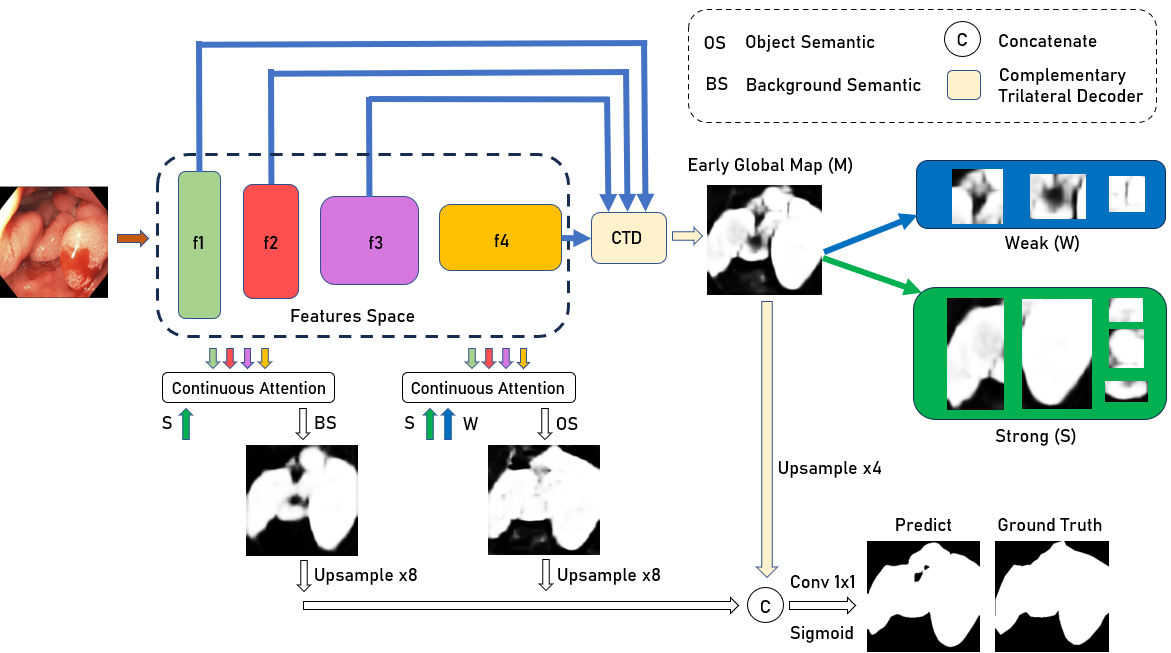
Abstract: Colonoscopy is a common and practical method for detecting and treating polyps. Segmenting polyps from colonoscopy image is useful for diagnosis and surgery progress. Nevertheless, achieving excellent segmentation performance is still difficult because of polyp characteristics like shape, color, condition, and obvious non-distinction from the surrounding context. This work presents a new novel architecture namely Adaptation of Distinct Semantics for Uncertain Areas in Polyp Segmentation (ADSNet), which modifies misclassified details and recovers weak features having the ability to vanish and not be detected at the final stage. The architecture consists of a complementary trilateral decoder to produce an early global map. A continuous attention module modifies semantics of high-level features to analyze two separate semantics of the early global map. The suggested method is experienced on polyp benchmarks in learning ability and generalization ability, experimental results demonstrate the great correction and recovery ability leading to better segmentation performance compared to the other state of the art in the polyp image segmentation task. Especially, the proposed architecture could be experimented flexibly for other CNN-based encoders, Transformer-based encoders, and decoder backbones.
pip install -r requirements.txtDownload the datasets from here and put them into polyp/ folder.
python train.py --epoch 200 --batch_sise 16 --lr 1e-4 --min_lr 1e-8 --val_name kvasir
python test.py --pretrain_path weight.hdf5 --test_name kvasir
You could download results of baseline methods from here.
If you have found our work useful, please consider citing:
@inproceedings{ADSNet,
author = {Quang Vinh Nguyen and Van Thong Huynh and Soo-Hyung Kim},
title = {Adaptation of Distinct Semantics for Uncertain Areas in Polyp Segmentation},
booktitle = {BMVC},
year = {2023}
}