D-SarcNet: A Dual-stream Deep Learning Framework for Automatic Analysis of Sarcomere Structures in Fluorescently Labeled hiPSC-CMs
Huyen Le, Khiet Dang, Nhung Nguyen, Mai Tran, and Hieu Pham*
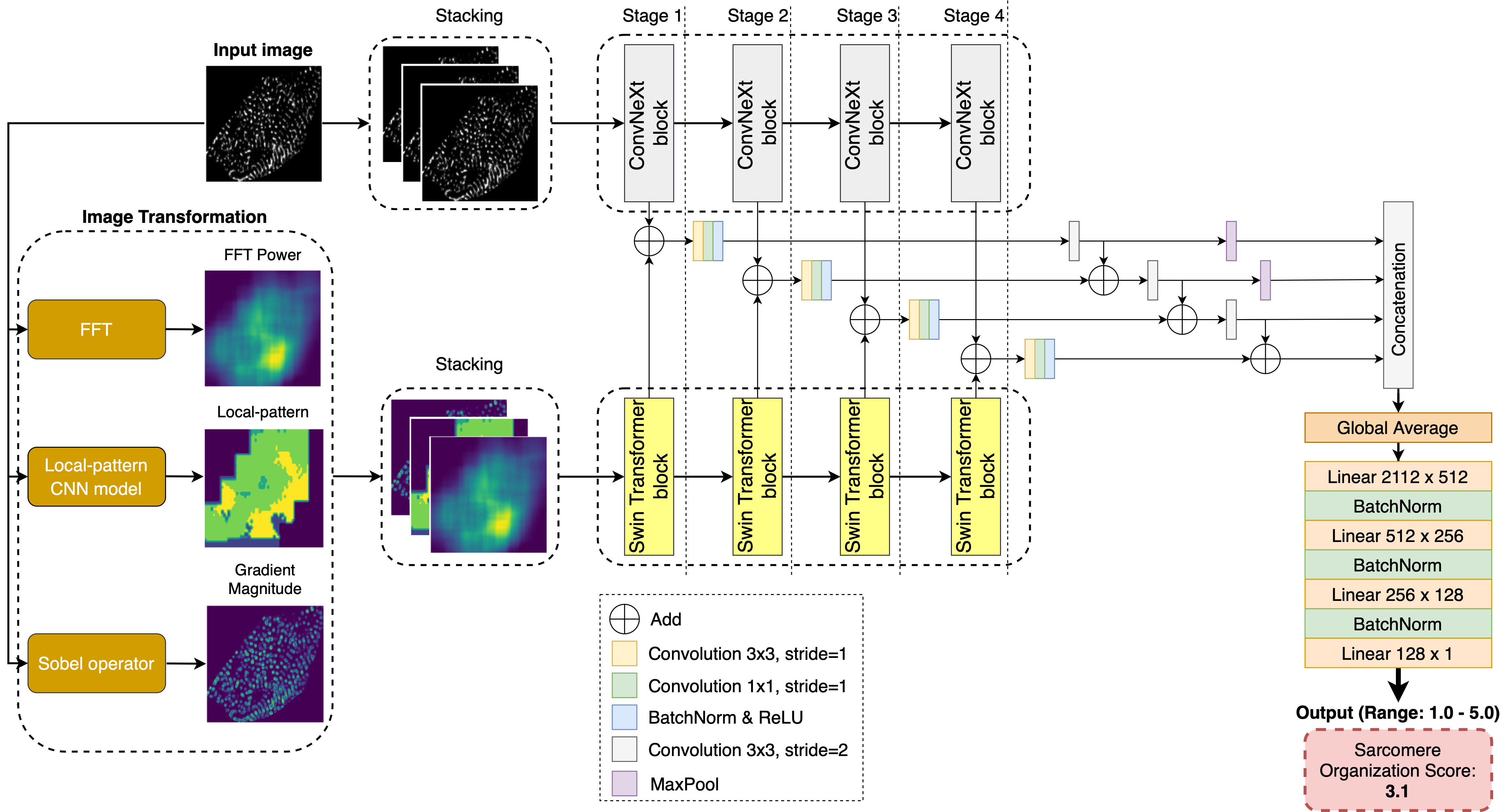
D-SarcNet is a dual-stream deep learning framework that enables high-throughput and accurate quantification of sarcomere structure organization in hiPSC-CMs single-cell images. Specifically, the framework processes fluorescently labeled hiPSC-CM single-cell images as input and generates a continuous value ranging from 1.0 to 5.0, indicating the level of sarcomere structural organization for each image. In addition, we propose three image-based representations: Fast Fourier Transform (FFT) Power image, local patterns, and gradient magnitude, to differentiate multiple patterns of
The data is available at https://open.quiltdata.com/b/allencell/packages/aics/integrated_transcriptomics_structural_organization_hipsc_cm. Please download:
- Data 1: 2d_autocontrasted_fields_and_single_cells_fish_1/rescale_2D_single_cell_tiff_path
- Data 2: 2d_autocontrasted_fields_and_single_cells_fish_2/rescale_2D_single_cell_tiff_path
- CSV 1: 2d_autocontrasted_single_cell_features_fish_1/features/a749d0e2_cp_features.csv
- CSV 2: 2d_autocontrasted_single_cell_features_fish_2/features/1493afe2_cp_features.csv
- File meta 1: automated_local_and_global_structure_fish_1/metadata.csv
- File meta 2: automated_local_and_global_structure_fish_2/metadata.csv
Run the command pip install -r requirements.txt to install the necessary libraries.
Please run the file train.py using the following command:
python train.py --csvpath1 YOUR_PATH_OF_CSV1 --csvpath2 YOUR_PATH_OF_CSV2 --metapath1 YOUR_PATH_OF_META1 --metapath2 YOUR_PATH_OF_META2 --datapath1 YOUR_PATH_OF_DATA1 --datapath2 YOUR_PATH_OF_DATA2
The outputs would include:
- best_model_corr.pt: model with the highest Spearman Correlation on the validation dataset
- best_model_loss.pt: model with the lowest validation loss
- best_model_r2.pt: model with the highest R2 score
- training.log: file log of the training and validation processes. In the end, the two best models (best_model_corr.pt and best_model_loss.pt) would be evaluated on the testing dataset on the four evaluation metrics of Spearman Correlation, MAE, MSE, and R2 score.
- loss.png: an image drawing training and validation loss through epochs
The output would include the results of the four evaluation metrics (Spearman Correlation, MAE, MSE, and R2 score) on the testing dataset.
Gerbin, K.A., Grancharova, T., Donovan-Maiye, R.M., Hendershott, M.C., Anderson, H.G., Brown, J.M., Chen, J., Dinh, S.Q., Gehring, J.L., Johnson, G.R., Lee, H., Nath, A., Nelson, A.M., Sluzewski, M.F., Viana, M.P., Yan, C., Zaunbrecher, R.J., Cordes Metzler, K.R., Gaudreault, N., Knijnenburg, T.A., Rafelski, S.M., Theriot, J.A., Gunawardane, R.N.: Cell states beyond transcriptomics: Integrating structural organization and gene expression in hipsc-derived cardiomyocytes. Cell Syst 12(6), 670–687 e10 (2021). DOI 10.1016/j.cels.2021.05.001. URL https://doi.org/10.1016/j.cels.2021.05.001
If you have any concerns or support on running this repository, please drop an email at huyen.lm@vinuni.edu.vn