This is a generic implementation of Variational Network (loop unrolling) for MR image reconstruction, proposed by Hammerick et al.. MRI data used to train the network is introduced and described by Johannes Schmidt and Sebastian Kozerke.
- Download 2D short axis cardiac MRI dataset https://polybox.ethz.ch/index.php/s/pYQc9w3M5MgRd4X
- Inspect configuration of the network in
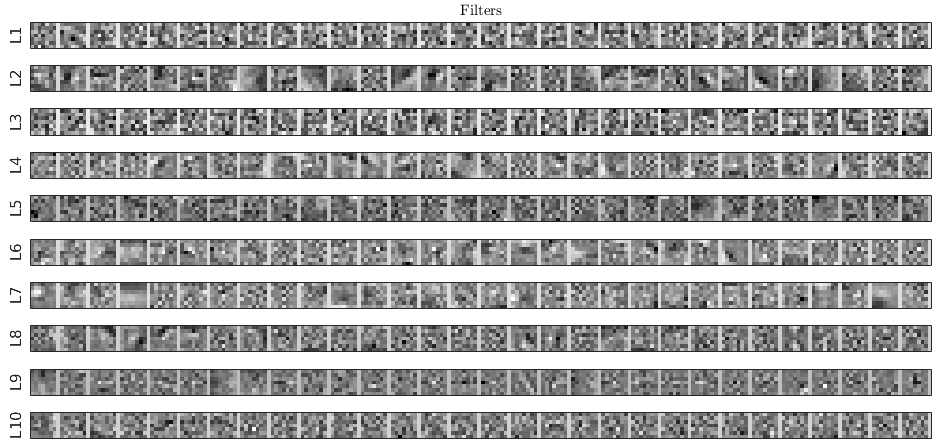
recon_model_300_066.py. We used:- 10 layers
- 7x7 filters
- 30 filters per layer
- 35 knots to parametrize activations
- cubic interpolation for activations
- Run
example.shfor training, or use saved parametersckpts/model.ckptand runrecon_model_300_066.py - Run matlab script
report_imgs.mfor illustrations of reconstruction and learned parameters
For training we used fully sampled 128x128 short axis MR images of the heart with artificially generated smooth phase offset.
The k-space was retrospectively undersampled to achieve acceleration factor of ~3.
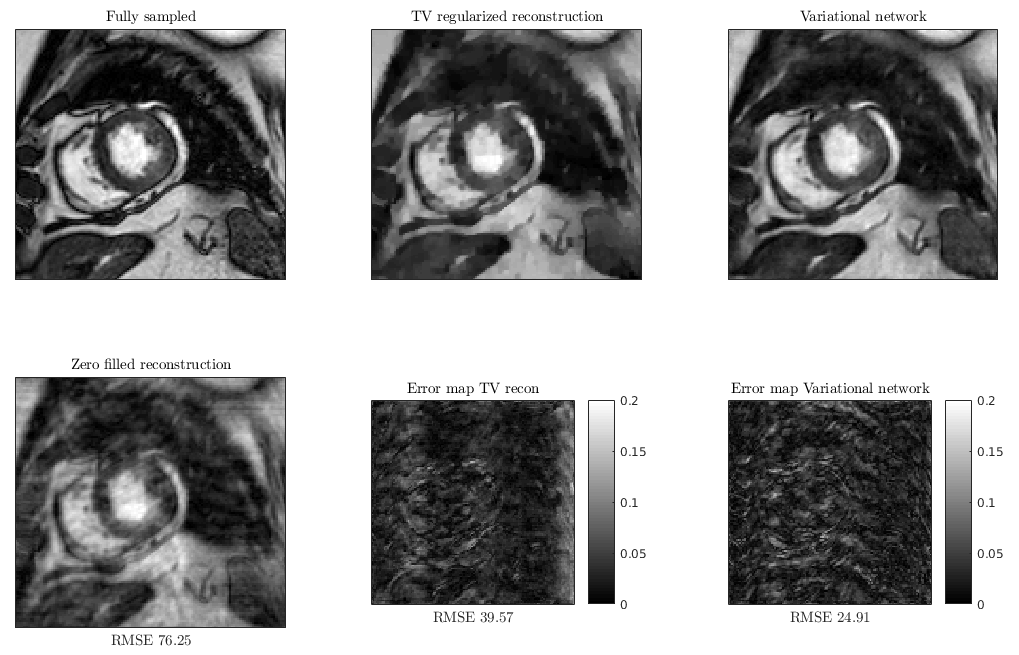
Variational Network reconstruction was compared to total variation (TV) regularized reconstruction (with the optimal regularization weight).
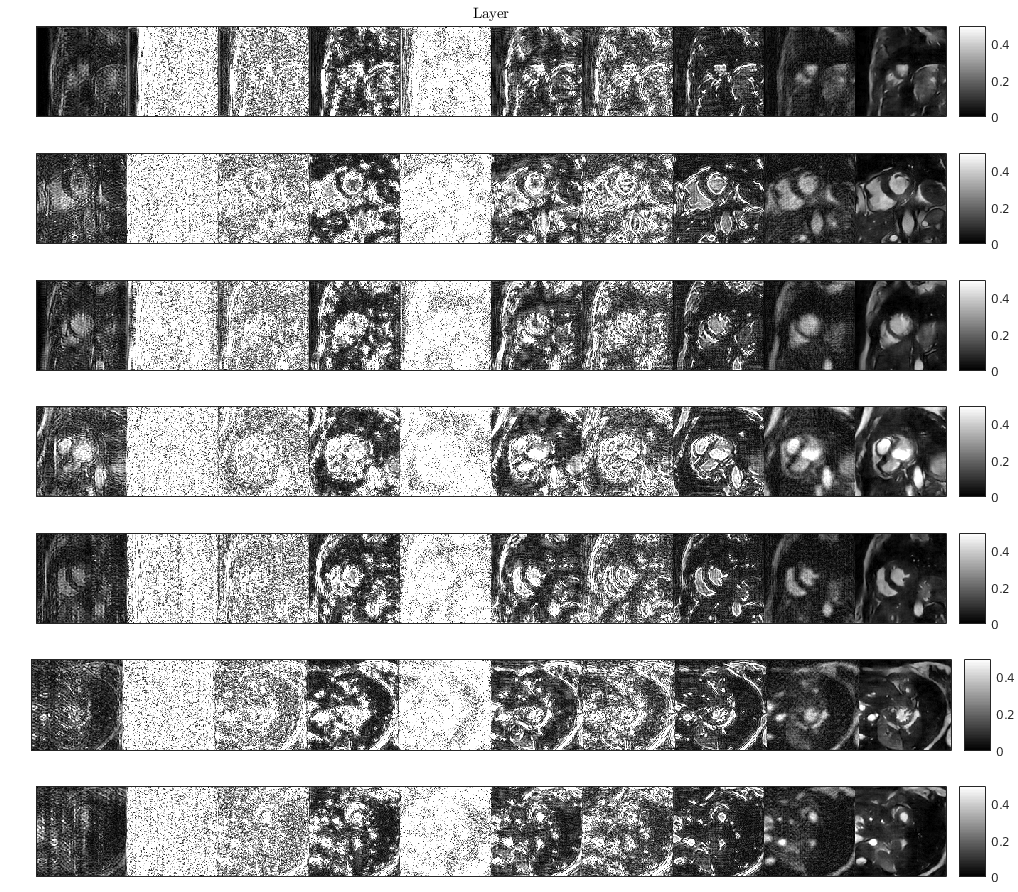
Reconstruction for each layer of the network:
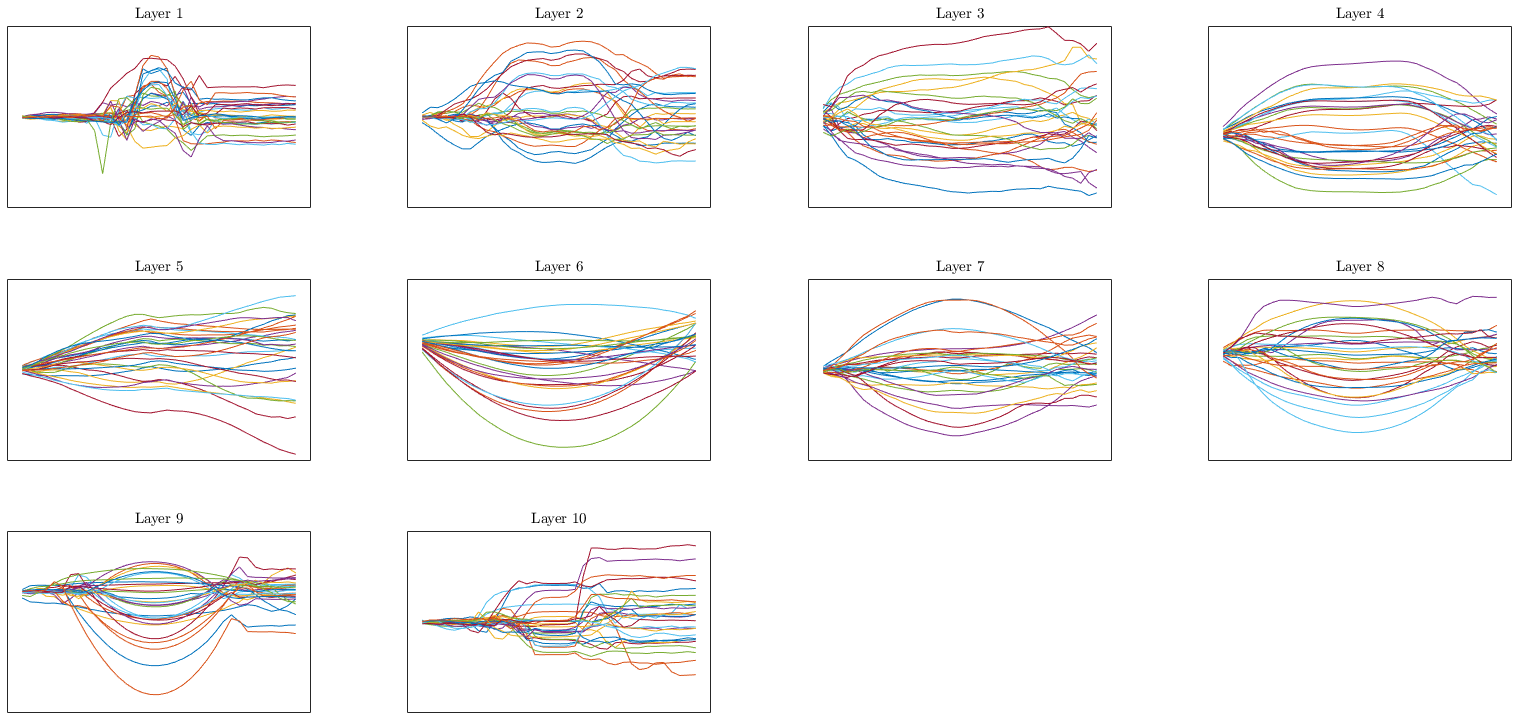
Learned activation functions (integral of activation, i.e. element-wise potentials):
- We are reconstructing smaller images compared to original work. Hence different filter sizes are used.
- Multi coil acquisition as well as coil sensitivity maps are not modelled.
- Batch size is fixed at configuration time.
- Cubic interpolation is used by default instead of RBF, since it is more memory efficient. You can choose
linear,cubic, orRBFin the network configuration. - Maximal estimated filter response is fixed for all layers.
The code is developed by Valery Vishnevskiy, Cardiac Magnetic Resonance group, Institute for Biomedical Engineering, ETH Zurich.