NB! We did not uploaded part of our data, because it will be used in our future work and published later as a part of article.
Flax wilt is an aggressive disease caused by soil-borne pathogen Fusarium oxysporum f. sp. lini (FOLINI). The fungus poses a major threat to flax production worldwide, as occasionally yield losses reach 70%. Here, we present first insights into regulatory mechanisms involved in response of flax varieties to the infection.
Our ain is to study the regulation of expression of genes responsible for the molecular mechanisms of flax response to FOLINI infestation
Objectives:
- RNA-seq and BS-seq sequencing data processing
- Alignment of reads to the reference genome
- Filtering and preprocessing data
- Statistical analysis of contrasts between various conditions of flax and FOLINI
- Differential gene expression analysis (DE)
- Determination of differential methylated DNA regions (DMR)
- Annotation enrichment analysis with terms:
- Gene Ontology
- Plant Reactome (for flax)
- Integrative Analysis (DE genes ✕ DM genes → Enrichment)
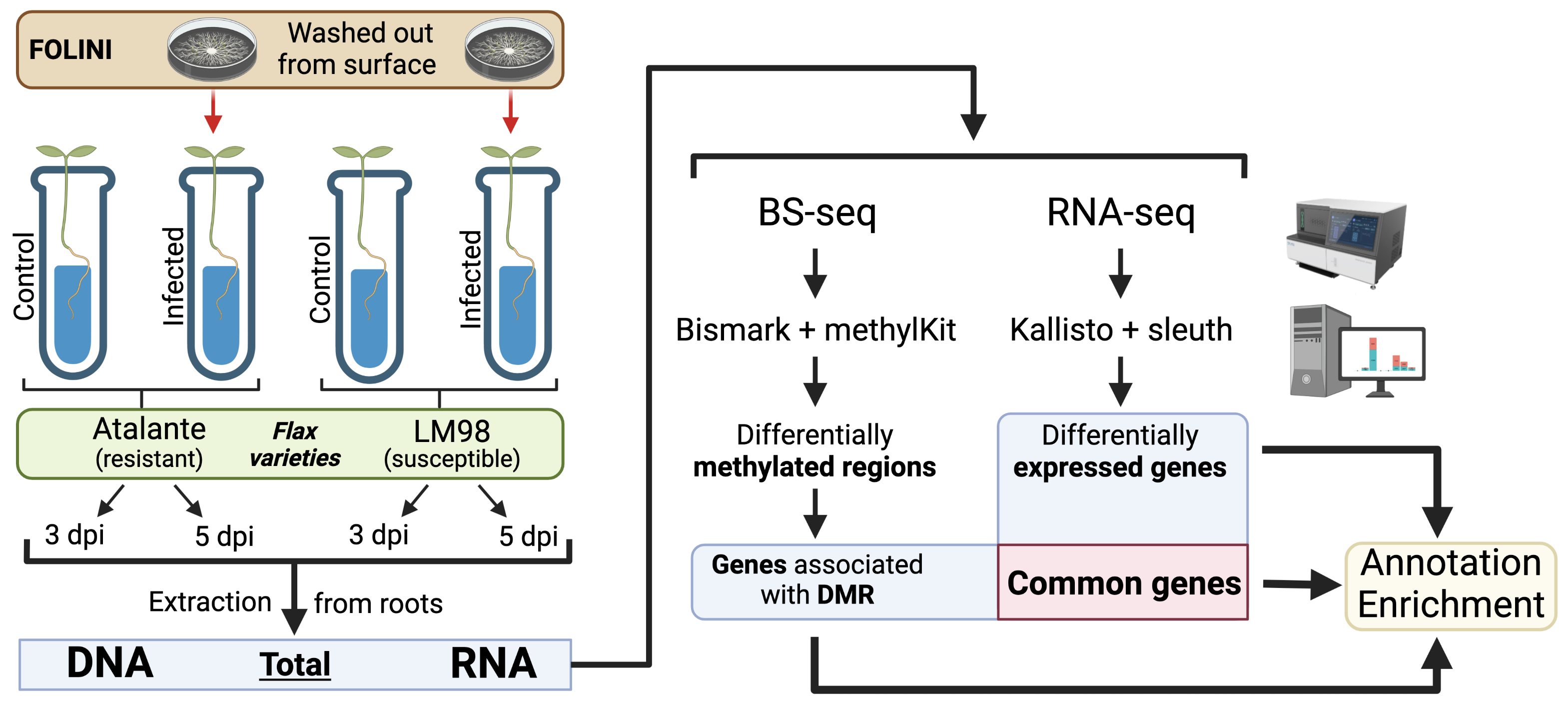
In this study we examined susceptible and resistant flax varieties, namely, LM98 and Atalante, infected by highly virulent MI39 FOLINI isolate. Uninfected plants and pure culture of the fungus served as controls. Flax samples were harvested on the third and the fifth day post inoculation (dpi). A total RNA and DNA mixtures (i.e., flax and fungus) was extracted from infected plant roots and sequenced. DNA was converted by bisulfite before sequencing.
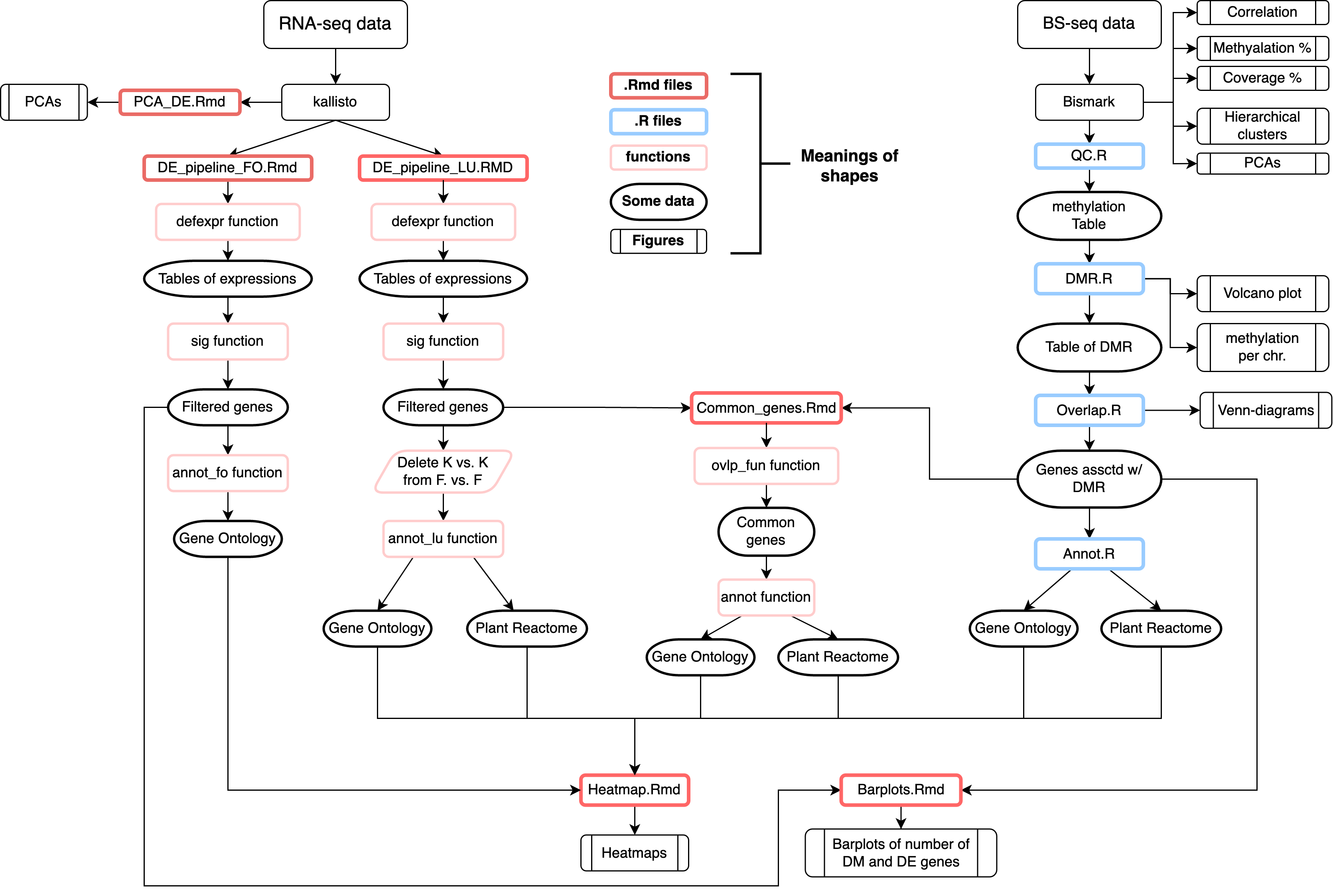
Reads were were aligned on reference genomes – MI39 isolate for fungus and flax genome assemble with accession number GCA_000224295.2. Aligning reads and abundances quantification of transcripts were obtained by processing RNA-seq data with Kallisto. Aligning of reads and identification of methylation positions and context (CpG, CHG, CHH, where H = A/C/T) were obtained by processing BS-seq data with Bismark. Quality control and filtering were complete with sleuth and methylKit R packages for transcriptome and methylome data respectively. Transcripts and methylation regions were filtered by number of cytosines and coverage - minimal coverage 5 (at least in half samples) for RNA-seq data and 10 for BS-seq data (default settings)
We planned to analyze 12 flax and 4 FOILINI comparisons between different experimental conditions in differential expression and differential methylation analyses.
| Difference between | |||
|---|---|---|---|
| LMF3 vs. FO | LMF5 vs. FO | AtF3 vs. FO | AtF5 vs. FO |
| Difference between | |||
|---|---|---|---|
| Between infected and control samples | During infection | ||
| LMF3 vs. LMK3 | LMF5 vs. LMK5 | LMF5 vs. LMF3 | |
| AtF3 vs. AtK3 | AtF5 vs. AtK5 | AtF5 vs. AtF3 | |
| Between varieties of flax | Between control samples | ||
| AtF3 vs. LMF3 | LMK5 vs. LMK3 | AtK5 vs. AtK3 | |
| AtF5 vs. LMF5 | AtK3 vs. LMK3 | AtK5 vs. LMK5 | |
Initial analysis consisted of checking of principal components, hierarchical clustering and correlation between samples. The determination of DE genes (DEG) was performed with the sleuth R package using adjusted p-value (Benjamini-Hochberg) and logarithm of fold change as thresholds. DMR were obtained by utilizing methylKit R package using adjusted p-value (Benjamini-Hochberg) and methylation percentage difference.
Integrative analysis: we intersected the coordinates of DMRs and genes and obtained a list of genes that were presumably regulated by methylation (common genes). All three lists of genes (DE, DM, common) were enriched by Gene Ontology and Plant reactome terms using XGR R package
| Dif.expression | Dif.methylation | Enrichment | |
|---|---|---|---|
| Adjusted p-value | 0.01 | 0.01 | 0.01 |
| Difference between samples | 1.25 (logarithmic) | 25 % | - |
| Min. number of genes with a term | - | - | 3 |
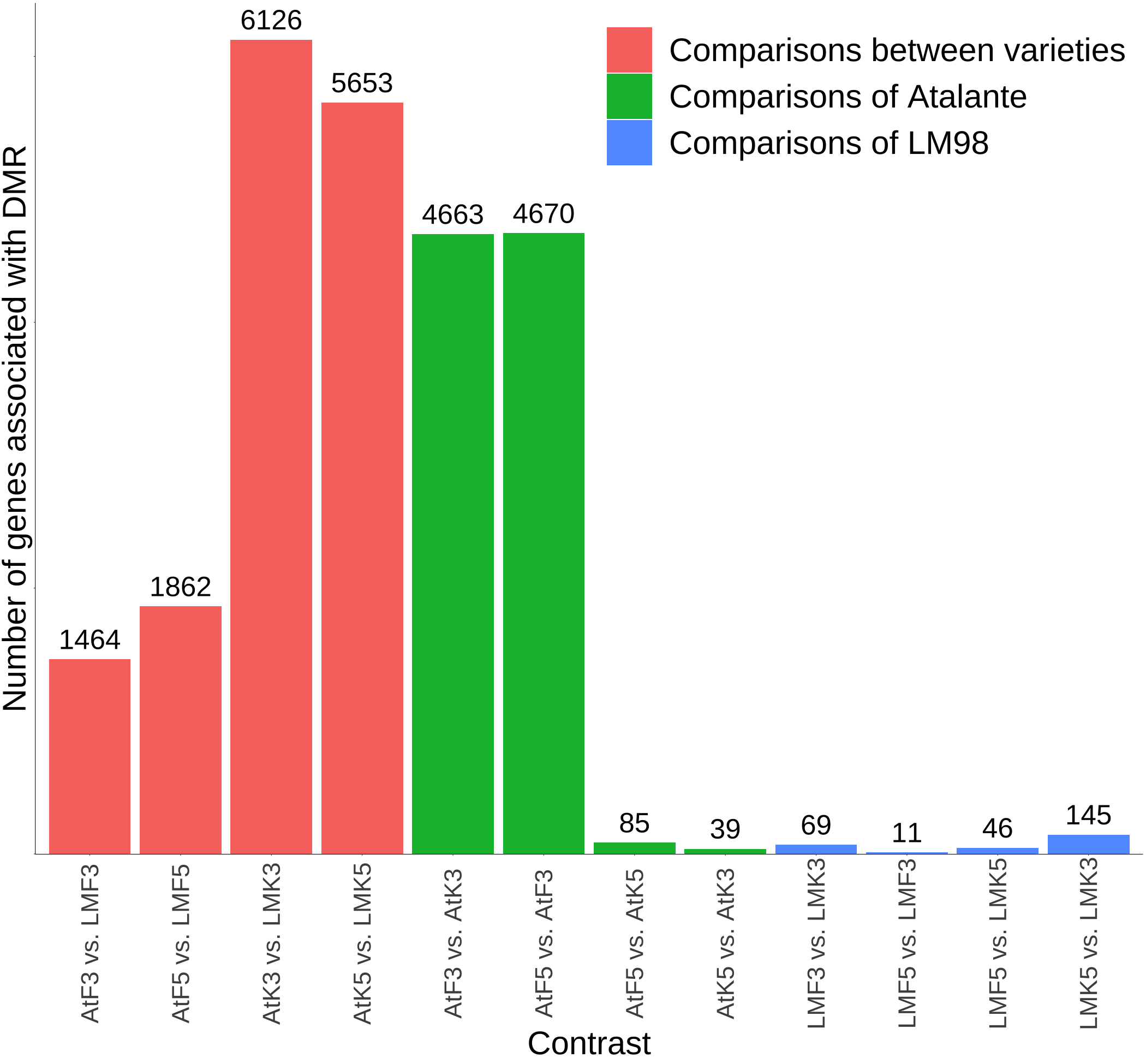
There were a lot of genes associated with DMR in infected Atalante on the 3rd dpi and their number decreased dramatically on the 5th dpi compared to control. Also, there were few DM genes between controls. Analysis of LM98 showed low number of DM genes between all conditions of the variety. Contrasts between LM98 and Atalante varieties showed large list of methylated genes in all contrasts, especially between control conditions. However, such big number of DMR discovered in these contrasts is the consequence of low methyaltion level in LM98 variety. FOLINI had not enough data about methylation to process
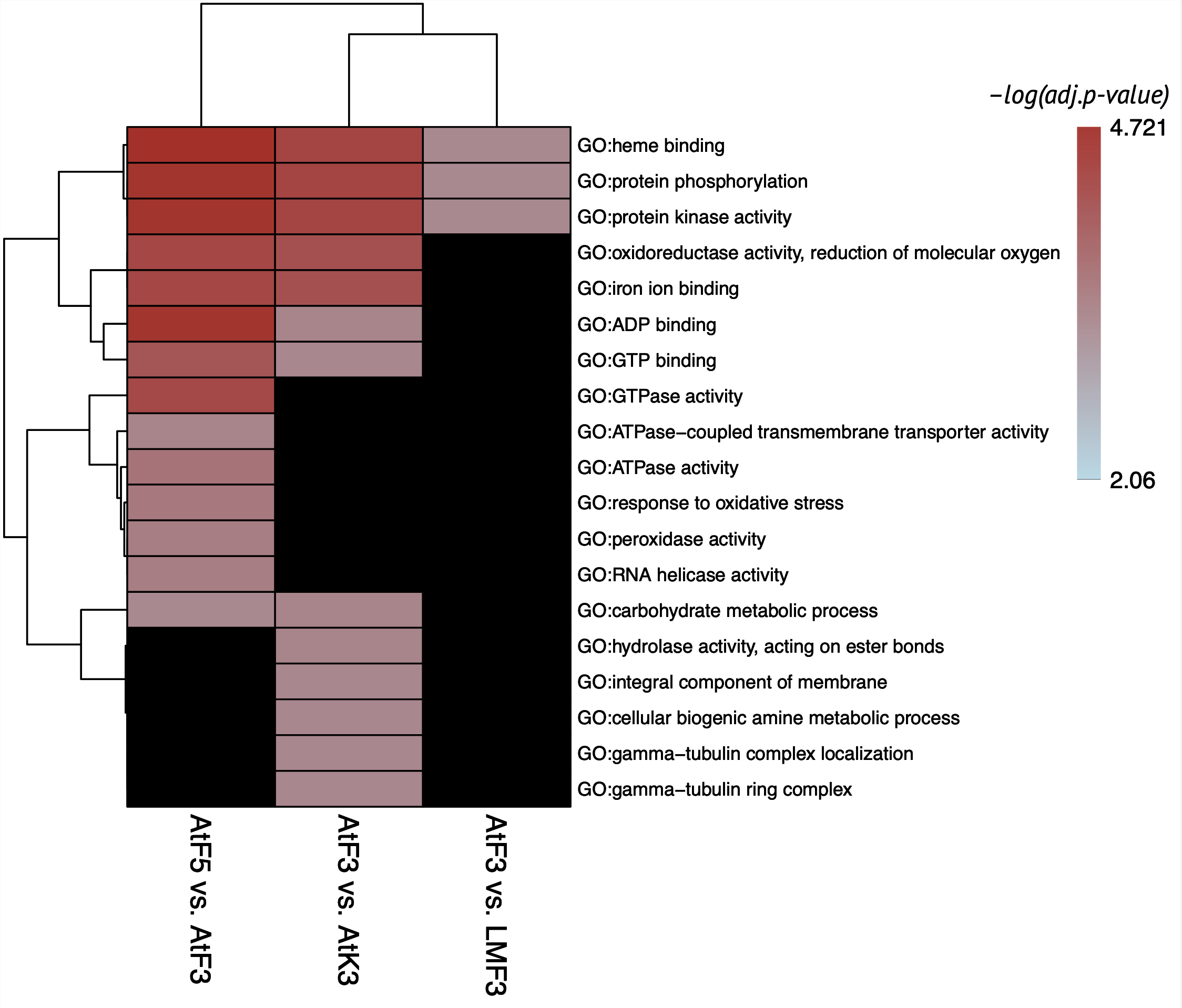
Enriched GO terms were found only for 3 comparisons. Contrasts of LM98 conditions did not enriched due to low number of obtained genes. Surprisingly, no terms were found for comparisons of control samples between varieties, although big number of genes associated with DMR were reported for these contrasts. Main enriched GO terms found AtF5 vs. AtF3 contrast connected with:
- Phosphorylation
- ATPase/GTPase activity
- Response to oxidative stress
- Carbohydrate metabolism
The important terms found AtF3 vs. AtK3 contrast associated with the same functions with addition of terms of carbohydrates metabolism.
Few enriched Plant Reactome terms were found. They denoted the synthesis of some protective agents (e.g. momilactones)
[!] The annotation enrichment analysis could not be performed with the genes from the differential methylation analysis for the LM98 variety conditions because of their small number. As the most important part of our study was integrative analysis, we decided to focus only on Atalante variety to study molecular mechanisms of resistance to fusariosis.
| Comparison | Number of DEG |
|---|---|
| AtF3 vs. AtK3 | 3790 |
| AtF5 vs. AtK5 | 0 |
| AtF5 vs. AtF3 | 1617 |
| AtK5 vs. AtK3 | 1147 |
| Comparison | Number of DEG |
|---|---|
| AtF3 vs. FO | 1731 |
| AtF5 vs. FO | 1612 |
FO - control Fusarium
There were a lot of DEG in infected Atalante on the 3rd dpi and their number decreased to zero on the 5th dpi сompared to control. Also, there were more genes differentially expressed in the comparison of control samples than genes detected in the differential methylation assay. Number of DEG discovered in contrasts of FOLINI was enough for further analysis.
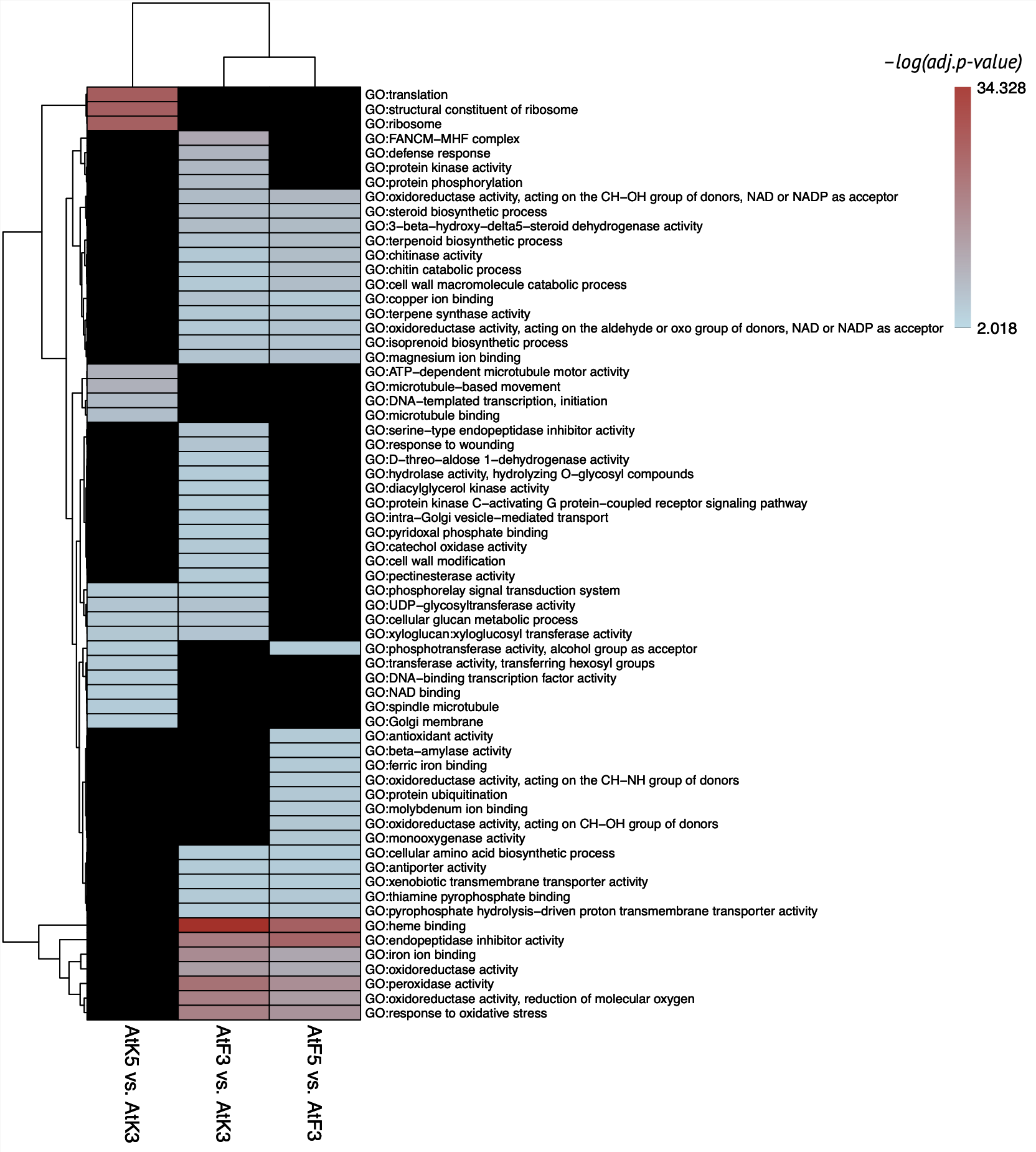
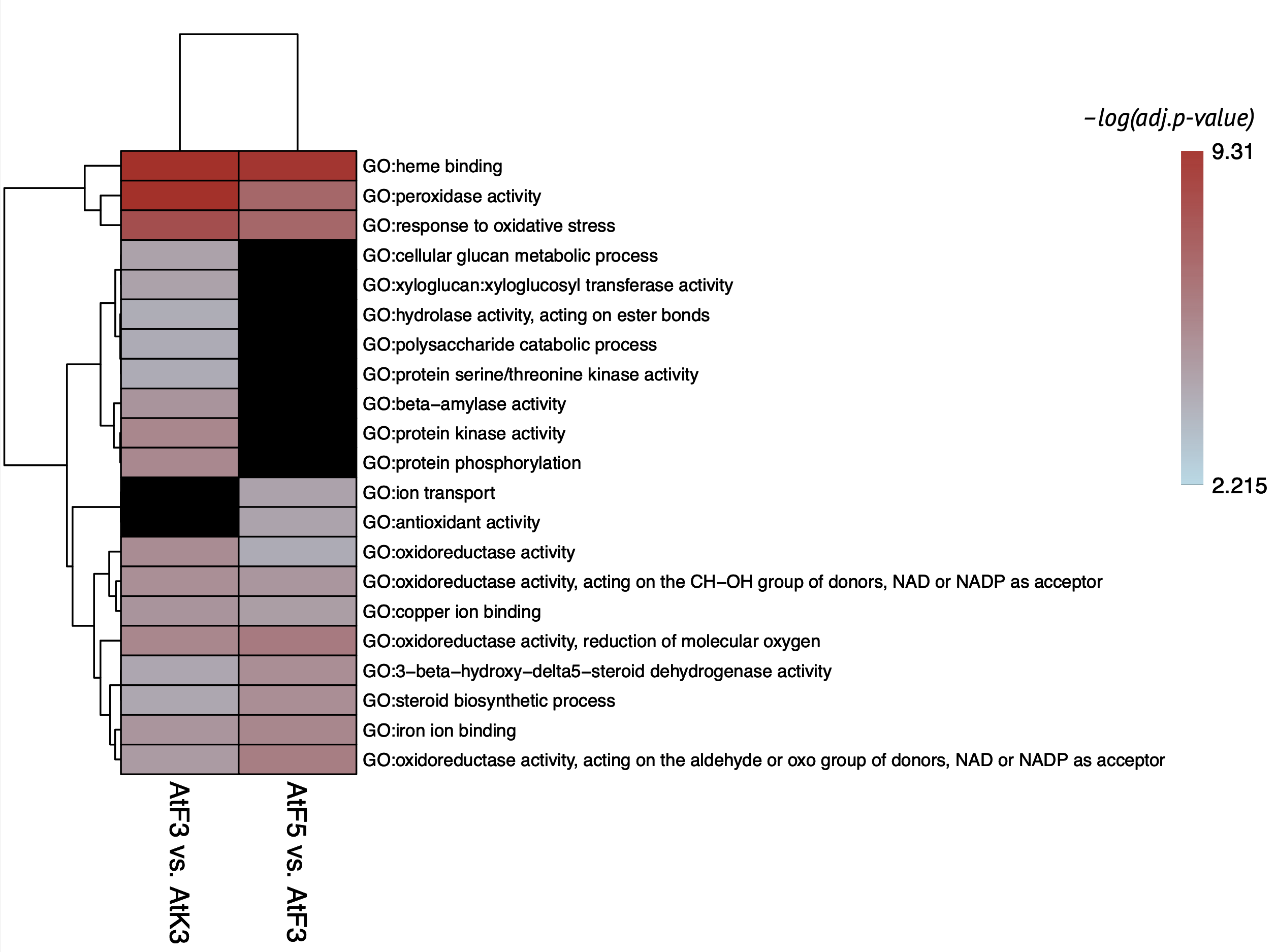
Atalante variety had strong specific response to infection on 3rd dpi. The main functional terms connected to defense response that faded down on 5th dpi:
- Phytohormones synthesis (brassinosteroids, terpenoids, jasmonic acid, ethylene, etc.)
- Response to oxidative stress (specific enzymes activity and ion binding)
- Cell wall modification
- Kinase activity
- Endopeptidase inhibitors
- Hydrolysis of polysaccharides
| Important enriched Plant Reactome terms |
|---|
| Ethylene synthesis and signaling |
| Momilactone synthesis |
| Jasmonic acid biosynthesis |
| Methylerythritol phosphate pathway |
| Recognition of fungal an pathogens and immunity response |
| Response to heaavy metals |
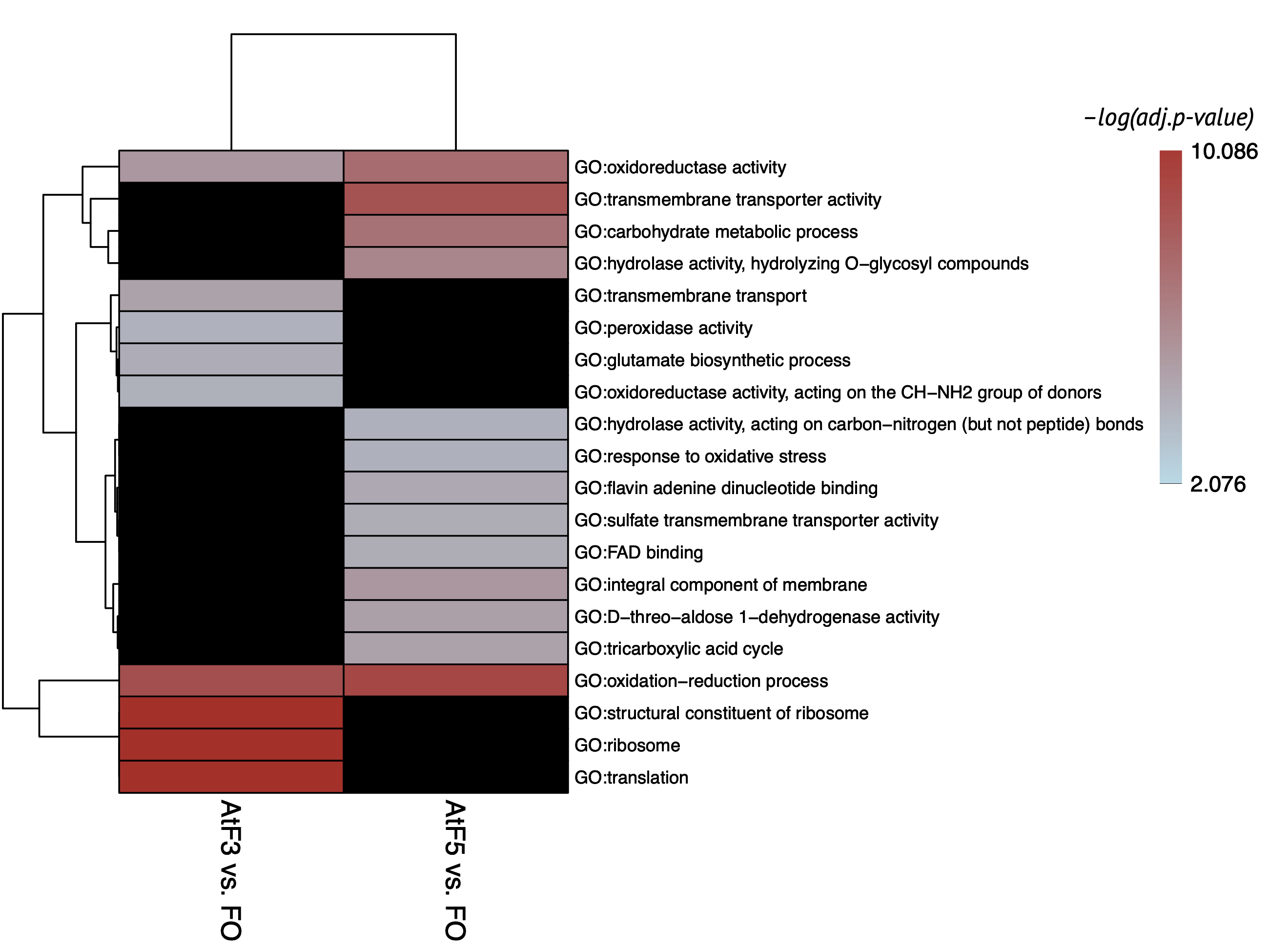
Enriched GO terms on 3rd and 5th dpi for FOLINI are markedly different.
For the 3rd day of infection discovered annotations were connected with:
- Synthesis of peptides
- Response to oxidative stress
On the 5th dpi terms were associated with:
- Transmembrane transport
- Carbohydrates metabolism
- Response to oxidative stress
After intersection of DEG and genes associated with DMR we found no common genes in AtF5 vs. AtK5 contrast and one gene in AtK5 vs. AtK3
There were enough common genes obtained for AtF3 vs. AtK3 and AtF5 vs. AtF3 for subsequent annotation enrichment analysis. The same terms found for these contrasts were connected with response to oxidative stress (specific enzymes activity and ion binding). Also, infected Atalante compared to control on the 3rd dpi differs with terms associated with polysaccharides metabolism and kinases activity
- Susceptible variety of flax LM98 had no methylation dynamic during the Fusarium infection and were excluded from analysis
- Resistant variety of flax Atalante had strong reaction to FOLINI invasion on the 3rd dpi that faded down on the 5th dpi. General enriched terms of DEG revealed several functions involved in the defense response:
| Phytoharmones synthesis | Signalling and regulation |
| Response to oxidative stress | Associated with hypersensivity reaction |
| Cell wall modification | Pathogen spreading prevention |
| Kinases activity | Molecular signaling |
| Endopeptidase inhibitors | Supression of FOLINI catabolic enzymes |
| Hydrolisis of polysacharides | Fungus cell wall degradation |
- Integrative and differential methylation analyses showed that some molecular mechanisms of defense response partly regulated epigenetically by direct DNA methylation. Such mechanisms marked as bold text in the table.
- DEG of FOLINI turned out to be associated with peptides synthesis (possibly some effectors and enzymes) and response to oxidative stress. On the 5th dpi DEG were enriched with functional terms of transmembrane transport, carbohydrates metabolism and response to oxidative stress, thus pathogen continued to actively parasitise. FOLINI methylation data were insufficient for analysis which is in consistent with recent studies about low methylation dynamic in fungi
- DE - All information about the directory can be seen in it
- DMR - All information about the directory can be seen in it
- Heatmaps - directory contains .png files of heatmaps of enriched GO and Plant Reactome terms
- Barplots - barplots of number of differentially expressed genes (DEG) and genes associated with differentially methyalted regions of DNA (DMR).
- Rmarkdown_scripts - .Rmd files which used to plot Heatmaps, Barplots and analyze common genes (DEG ∩ genes associated with DMR)
- RDS_files - files that were created during the work of pipeline for fast access to varuiables from different scripts. This directory is empty as we cannot publish our full data.