Example pipeline with Nextflow used to assess results, comparing the metrics being computed with this workflow with TCGA results.
Nextflow workflows following the same structure as this repo's example will be integratable into the OpenEBench Virtual Research Environment (VRE).
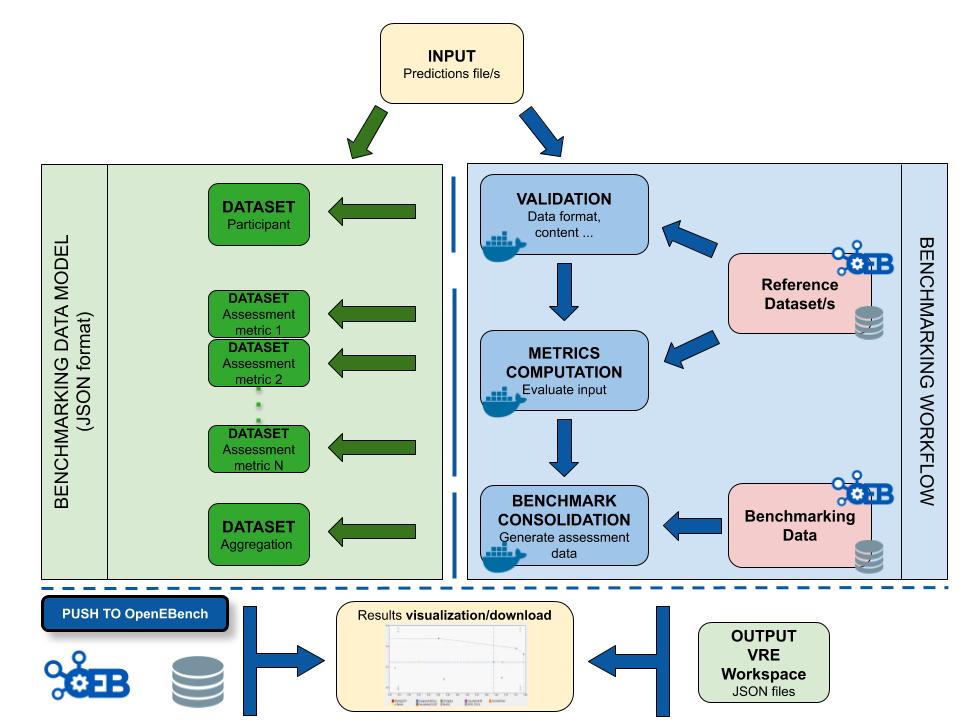
The workflow takes an input file with Cancer Driver Genes predictions (i.e. the results provided by a participant), computes a set of metrics, and compares them against the data currently stored in OpenEBench within the TCGA community. Two assessment metrics are provided for that predicitions. Also, some plots (which are optional) that allow to visualize the performance of the tool are generated. The workflow consists in three standard steps, defined by OpenEBench. The tools needed to run these steps must be in one or more Docker images, generated from Docker containers. Separated instances are spawned from these images for each step:
- Validation: the input file format is checked and, if required, the content of the file is validated (e.g check whether the submitted gene IDs exist)
- Metrics Computation: the predictions are compared with the 'Gold Standards' provided by the community, which results in two performance metrics - precision (Positive Predictive Value) and recall(True Positive Rate).
- Results Consolidation: the benchmark itself is performed by merging the tool metrics with the rest of TCGA data. The results are provided in JSON format and SVG format (scatter plot).
- TCGA_sample_data folder contains all the reference data required by the steps. It is derived from the manuscript:
Comprehensive Characterization of Cancer Driver Genes and Mutations, Bailey et al, 2018, Cell
- sample_out folder contains an example output for a worklow run, with two cancer types / challenges selected (ACC, BRCA)
In order to use the workflow you need to:
- Install Nextflow
- Clone this repository in a separate directory, and build locally the three Docker images found in it, running the
build.shscript within that repo. - Run it just using
nextflow run main.nf. Arguments specifications:
Usage:
Run the pipeline with default parameters:
nextflow run main.nf
Run with user parameters:
nextflow run main.nf --input {driver.genes.file} --public_ref_dir {validation.reference.file} --participant_id {tool.name} --goldstandard_dir {gold.standards.dir} --cancer_types {analyzed.cancer.types} --assess_dir {benchmark.data.dir} --results_dir {output.dir}
Mandatory arguments:
--input List of cancer genes prediction
--community_id Name or OEB permanent ID for the benchmarking community
--public_ref_dir Directory with list of cancer genes used to validate the predictions
--participant_id Name of the tool used for prediction
--goldstandard_dir Dir that contains metrics reference datasets for all cancer types
--event_id List of types of cancer selected by the user, separated by spaces
--assess_dir Dir where the data for the benchmark are stored
Other options:
--validation_result The output directory where the results from validation step will be saved
--assessment_results The output directory where the results from the computed metrics step will be saved
--outdir The output directory where the consolidation of the benchmark will be saved
--statsdir The output directory with nextflow statistics
--data_model_export_dir The output dir where json file with benchmarking data model contents will be saved
--otherdir The output directory where custom results will be saved (no directory inside)
Flags:
--help Display this message
Default input parameters and Docker images to use for each step can be specified in the config file. NOTE: In order to make your workflow compatible with the OpenEBench VRE Nextflow Executor, please make sure to use the same parameter names in your workflow.