MetaFast (METAgenome FAST analysis toolkit) is a toolkit for calculating a number of statistics of metagenome sequences and building the distance matrix between them.
Authors:
- Software: Artem Ivanov, Sergey Kazakov and Vladimir Ulyantsev,
ITMO University, Saint-Petersburg, Russia. - Testing: Veronika Dubinkina and Alexandr Tyakht,
SRI of Physical-Chemical Medicine, Moscow, Russia. - Idea, supervisor: Dmitry Alexeev,
SRI of Physical-Chemical Medicine, Moscow, Russia.
MetaFast documentation is available on the GitHub wiki page.
Here is a short version of it.
To run MetaFast you need to have JRE 1.6 or higher installed and only one script – metafast.sh, metafast.bat or metafast.jar.
You can download it from the last stable release in the GitHub 'Releases' section.
- For Linux and Mac OS: download
metafast.sh, run the commandchmod a+x metafast.sh, then run./metafast.shfrom the command line. - For Windows: download
metafast.batand run it from the command line. - For other OS: download
metafast.jarand run it via commandjava -jar metafast.jar.
Alternatively, you can build the newest version of the MetaFast from the repository:
git clone https://github.com/ctlab/metafast.git
cd metafast
ant
./out/metafast.sh --version
A new version of MetaFast software is being prepared for the release. New pipelines for comparative metagenomics data analysis have been implemented. Three recommended use cases (including the original one) and a detailed description of available tools are presented in Pipelines.md
To run MetaFast use the following syntax:
metafast.sh [<Launch options>] [<Input parameters>]metafast.bat [<Launch options>] [<Input parameters>]java -jar metafast.jar [<Launch options>] [<Input parameters>]
To view help for launch options and input parameters run metafast.sh --help or metafast.sh --help-all.
By running MetaFast a working directory is created (by default ./workDir/).
All intermidiate files, log file and final results are saved in it.
File output_description.txt is created after every run in the current and working directories.
It contains the description of every output file produced by the MetaFast.
Metafast run script also allows you to run subtools of whole process or different tools, that was included into the package.
To see the list of available additional tools, run metafast.sh --tools.
Download meta_test_1.fa, meta_test_2.fa and meta_test_3.fa and run the command:
./metafast.sh -i meta_test_1.fa meta_test_2.fa meta_test_3.fa
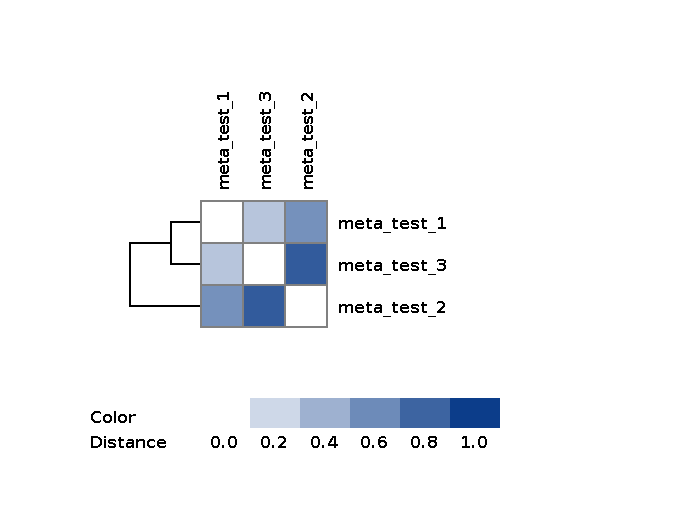
After it has finished, a distance matrix can be found in workDir/matrices/dist_matrix_<date>_<time>_original_order.txt:
# meta_test_1 meta_test_2 meta_test_3
meta_test_1 0.0000 0.5691 0.2981
meta_test_2 0.5691 0.0000 0.8448
meta_test_3 0.2981 0.8448 0.0000
The element matrix[i][j] is a distance between sample i and sample j.
K-mers frequency statistics is saved in workDir/kmer-counter-many/stats/<in-file>.stat.txt;
image file with heatmap and dendrogram is saved in workDir/matrices/dist_matrix_<date>_<time>_heatmap.png:
Q Does MetaFast works with paired-end reads?
A No, MetaFast accepts only one file per sample. If you have two files with paired-end reads (i.e. reads_1.fastq and reads_2.fastq), you need to combine them into one file before running MetaFast. On Unix-like systems, one can do it running cat reads_1.fastq reads_2.fastq > reads_all.fastq
Q Can I compare samples with different read lengths or the number of reads?
A Yes, you can do this without any specific preprocessing. MetaFast uses k-mers for comparing metagenomic sequences, so it will automatically normalize all values by the total number of k-mers per sample.
Q Can I compare metagenomes obtained from different sequencing platforms (i.e. Illumina vs 454-seq)
A Yes, you can. MetaFast extract features from each metagenome independently, so you can compare samples from different sequencing platforms.
If you use MetaFast in your research, please cite the following publication:
Ulyantsev V.I., Kazakov S.V., Dubinkina V.B., Tyakht A.V. & Alexeev D.G. (2016). MetaFast: fast reference-free graph-based comparison of shotgun metagenomic data. Bioinformatics, 32(18), 2760-2767. doi: 10.1093/bioinformatics/btw312
Please report any problems directly to the GitHub issue tracker.
Also, you can send your feedback to abivanov@itmo.ru.
The MIT License (MIT)