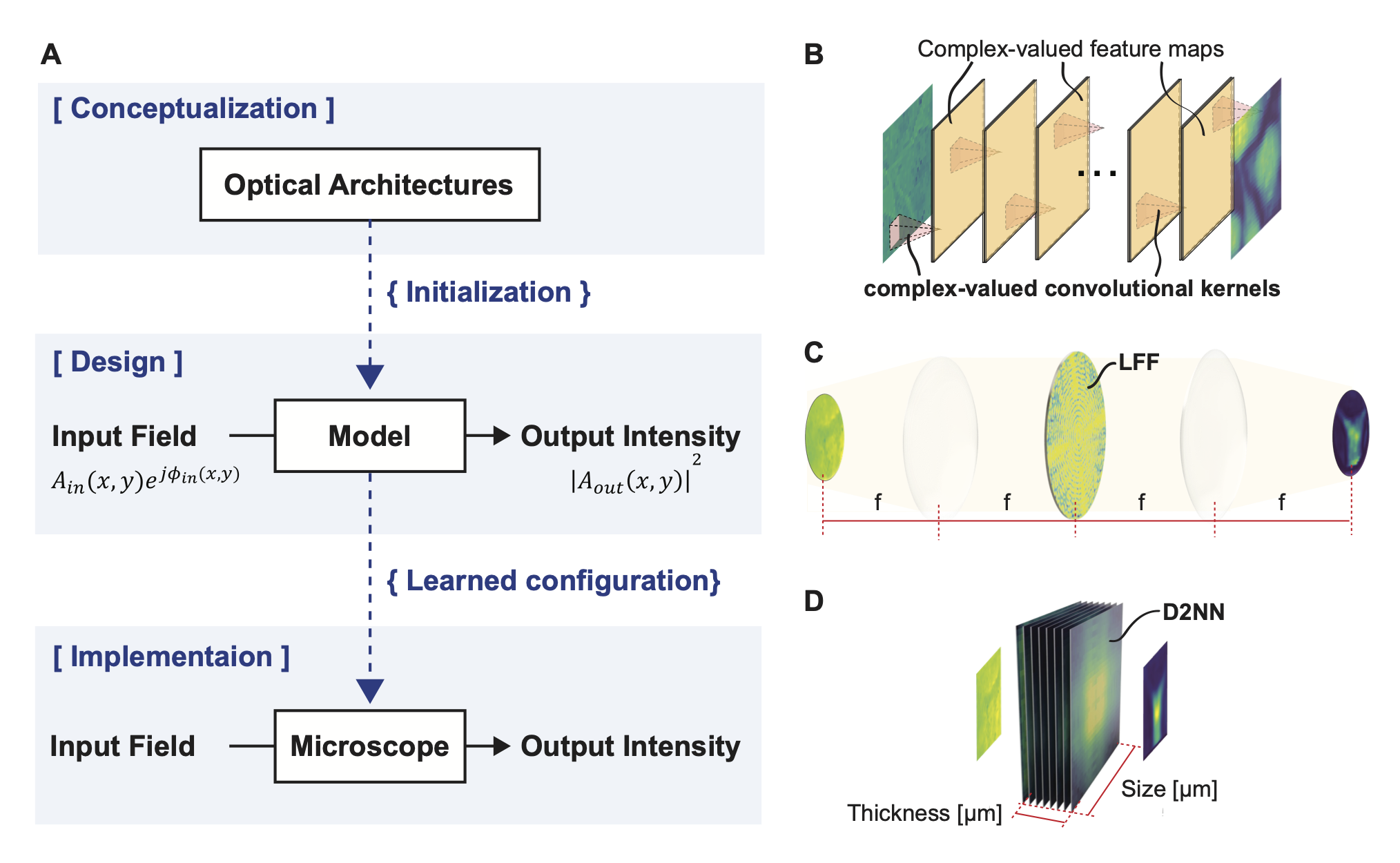
We introduce Differentiable Microscopy (
Here we provide the Google Colab notebooks to reproduce the results of our best performing models for each dataset for our microscopy designs.
-
Expected results of each notebook :
- SSIM score for the unseen dataset. SSIM values should match the below table.
- Groundruth and the corresponding reconstructed images will be printed in the notebooks
-
Expected runtime for Inference (for LFFs) on a normal desktop (CPU: Four 11th Gen Intel(R) Core(TM) i7-1165G7 (2.8GHz) cores) with GPU (2GB NVIDIA GeForce MX330) : 10.9785
$\pm$ 0.354 ms
| Method | MNIST [0, Pi] | MNIST [0, 2Pi] | HeLa [0,Pi] | HeLa [0,2Pi] | Bacteria [0, Pi] | Colab Notebook |
|---|---|---|---|---|---|---|
| Complex-valued linear CNN | 0.9982 | 0.7913 | 0.9417 | 0.8619 | 0.9938 |
git clone https://github.com/Bantami/All-Optical-QPM.gitHeLa dataset link, Bacteria dataset link
To download the dataset through the commandline, follow the below steps
pip install gdown
gdown https://drive.google.com/uc?id=10mj-mPmeStdOWvZnI-nVx8J89Wyp8RGj
gdown https://drive.google.com/uc?id=12AdUSF7DawnqVJMzfPzqD7mUa7kmZT2L
mkdir datasets/
unzip -qq hela.zip -d datasets/
unzip -qq bacteria.zip -d datasets/
Update dataloaders.py to set,
- MNIST
data_dir(any existing path will work) in line 30 and - HeLa
data_dirin line 60 - Bacteria
data_dirin line 89
- This should take only few minutes
## create new environment
conda create -n qpm_env python=3.6
source activate qpm_env
## Adding new environment to JupyterLab
conda install -c anaconda ipykernel -y
python -m ipykernel install --user --name=qpm_env
conda install pytorch==1.8.0 torchvision==0.9.0 torchaudio==0.8.0 cudatoolkit=11.1 -c pytorch -c nvidia
conda install -c conda-forge matplotlib
conda install -c conda-forge wandb
#install remaining packages through pip
pip install -r requirements.txtPlease find the training notebooks for each of the models for each dataset below.
run notebook_name.ipynb after selecting the newly added kernal (qpm_env)
results will be saved in the folder "results" which will be created in the parent directory w.r.t to where the notebook is located.
- Expected results of each training notebook (in
resultsfolder):- Saved model for latest epoch
- Loss curves figure
- Input/Reconstructed images comparison figure for each epoch (SSIM, L1, BerHu loss will be displayed)
- PhaseD2NN - MNIST [0,Pi]
- PhaseD2NN - MNIST [0,2Pi]
- PhaseD2NN - HeLa [0,Pi]
- PhaseD2NN - HeLa [0,2Pi]
- PhaseD2NN - Bacteria
- Complex-CNN - MNIST [0,Pi]
- Complex-CNN - MNIST [0,2Pi]
- Complex-CNN - HeLa [0,Pi]
- Complex-CNN - HeLa [0,2Pi]
- Complex-CNN - Bacteria
├── Colab
│ ├── cnn_inference_colab.ipynb
│ ├── d2nn_pretrained_model_inference_colab.ipynb
│ ├── GPC_baseline_inference_colab.ipynb
│ └── lff_pretrained_model_inference_colab.ipynb
│ └── PhaseD2NN_pretrained_model_inference_colab.ipynb
├── colab_setup.sh
├── modules
│ ├── d2nn_layers.py
│ ├── d2nn_models.py
│ ├── dataloaders.py
│ ├── datasets.py
│ ├── diffraction.py
│ ├── eval_metrics.py
│ ├── fourier_model.py
│ ├── loss.py
│ ├── other_models.py
│ ├── train.py
│ ├── train_utils.py
│ └── vis_utils.py
├── Notebooks
│ ├── ComplexCNN
│ │ ├── complexCNN_HeLA.ipynb
│ │ ├── complexCNN_HeLA_pi.ipynb
│ │ └── complexCNN_MNIST.ipynb
│ │ └── complexCNN_MNIST_2pi.ipynb
│ │ └── complexCNN_Bacteria.ipynb
│ ├── GPC.ipynb
│ ├── LearnableFourierFilter
│ │ ├── LFF_HeLA.ipynb
│ │ ├── LFF_HeLA_pi.ipynb
│ │ └── LFF_MNIST.ipynb
│ │ └── LFF_MNIST_2pi.ipynb
│ │ └── LFF_Bacteria.ipynb
│ ├── PhaseD2NN
│ │ ├── PhaseD2NN_hela_2pi.ipynb
│ │ ├── PhaseD2NN_hela_pi.ipynb
│ │ └── PhaseD2NN_mnist.ipynb
│ │ └── PhaseD2NN_mnist_2pi.ipynb
│ │ └── PhaseD2NN_bacteria.ipynb
│ └── results
├── overview.png
├── README.md
└── requirements.txt