This is a repository for calculating the Fisher-Rao Distance between densities and performing hypothesis testing.
In addition to the fact that the Fisher-Rao Distance is a proper distace metric, key advantages of using the Fisher-Rao Distance for comparing sets of data are that it can be used:
- parametrically or non-parametrically
- for scalar or multi-dimensional data
- in various domains (i.e. Rn or Sn ) by changing the form of the density
In this current implementation, we use a nonparametric Gaussian KDE and LOOCV for calculating its bandwidth parameter.
For this example we generate and compare three data sets generated from a Normal Distribution in R10 and mapped to R.
using FisherRaoDistance
using KernelDensityEstimatePlotting #currently not working in Atom; needs Atom dev update
The density estimation requires sets of points. These points can either be the original data or can be the result of some sort of dimension reduction. This example calculates pairwise distances and uses those in a classicalMDS setting
points1 = randn(10, 500)
points2 = randn(10, 500)
points3 = randn(10, 500) + ones(1, 500) * 1.5
#below calculates the pairwise Euclidean distance matrix between the points
#and uses those for classical mds
Points = [points1, points2, points3]
lowdimpoints = get_low_dim_points(Points, 1)
pdf1 = kde!(lowdimpoints[:,:,1])
pdf2 = kde!(lowdimpoints[:,:,2])
pdf3 = kde!(lowdimpoints[:,:,3])
#plot pane is not currently working in Atom-- waiting for Atom dev fix
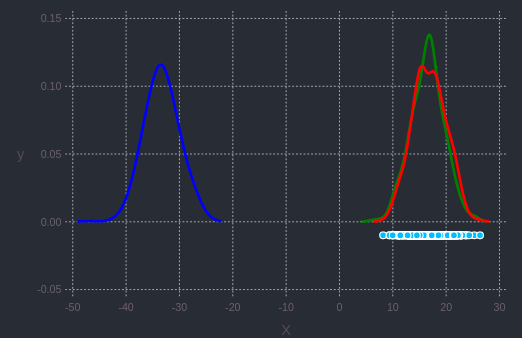
plot([pdf1; pdf2; pdf3], c = ["red"; "green"; "blue"])
dfr1_2 = fisherraodistance(
pdf1,
pdf2,
lowdimpoints[:, :, 1],
lowdimpoints[:, :, 2],
)
returns: 0.089
dfr1_3 = fisherraodistance(
pdf1,
pdf3,
lowdimpoints[:, :, 1],
lowdimpoints[:, :, 3],
)
returns: 0.495
p1_2 = fisherraotest(pdf1, pdf2, n1, n2, dfr1_2)
returns: 0.62
p1_3 = fisherraotest(pdf1, pdf3, n1, n3, dfr1_3)
returns: 0.00
data = rand(100) #generate some random data
pdf = kde!(data) #estimate a pdf from given data
data_likelihoods = evaluateDualTree(pdf,data) #get data likelihoods
sample_points, ind = sample(pdf,100) #sample 100 points from the pdf
Henning, Wade. "A Framework for Comparing Shape Distributions", 2014, Ph.D. Thesis, Florida State University.
Henning, Wade; Srivastava, Anuj. "A Two-Sample Test for Statistical Comparisons of Shape Populations", 2016, IEEE Winter Conference on Applications in Computer Vision
Srivastava, Anuj; Jermyn, Ian; Joshi, Shantau. "Riemannian analysis of probability density functions with applications in vision", 2007, IEEE Computer Vision and Pattern Recognition
Sudderth, Erik B.; Ihler, Alexander; et al. "Nonparametric belief propagation.", 2010, Communications of the ACM