This repository includes codes and data used in the "Single-cell spatial immune landscapes of primary and metastatic brain tumors" paper.
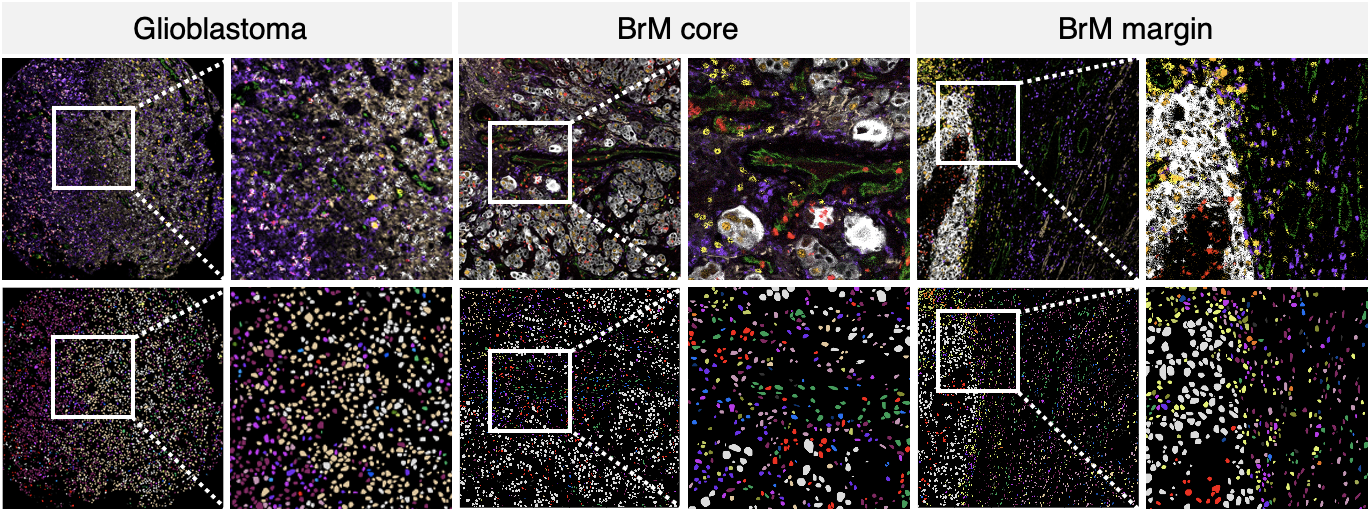
The emergence of single cell technologies in recent years has enabled characterization of the tumor microenvironment at unprecedented depth and has highlighted an incredible degree of cellular diversity amongst tumor cells and their niche. Anti-tumor immunity relies on coordinated spatially defined cell-cell relationships within the tumor microenvironment 1,2 , yet many single cell studies lack spatial context by sequencing dissociated tissues 3 . This has become increasingly critical for tumor types that are highly influenced by unique immune niches, such as the myeloid- rich lymphoid-cold microenvironment of brain tumors 4 . Here we apply imaging mass cytometry to characterize the immunological landscape of 140 glioblastoma and 46 brain metastasis patient tumor samples, with clinical data available for all patients. Single cell analysis of over 1.1M cells across 389 high-dimensional histopathology images enabled spatial resolution of immune lineages and activation states, revealing differences in immune landscapes between primary glioblastoma and brain metastases from diverse solid cancers. These analyses also identified cellular neighbourhoods associated with survival in patients with glioblastoma, which were leveraged to identify a unique population of recruited MPO+ macrophages that had an unexpected association with long-term survival. Our findings add depth towards understanding the putative regulatory impact of monocyte-derived macrophages and tissue-resident microglia on brain tumor progression and build context around how these cell types interact with their niche. Our study serves as a new resource for primary brain tumors and metastases, reinforcing the value of integrating spatial resolution into single cell datasets to improve our understanding of the microenvironmental contexture of cancer.
You can find the detail of each analysis in the following repositories.
The data folders on Zenodo are structured as follows:
- BRAIN_IMC_MaskTif: this folder contains masked multiplex images of the IMC cores.
- BRAIN_IMC_Segmentation: this folder containes the nuclei segmentation information.
- BRAIN_IMC_CellType: this folder containes the cell type information for each of the IMC cores.
- Clinical Data GBM BrM.xlsx: this folder containes clinical data with regard to the images listed above.