This is a Keras with TensorFlow backend implementation for ISIC 2019 Challenge Task 1: classify dermoscopic images among nine different diagnostic categories without meta-data. The proposed approaches implemented in this repository were ranked 25th out of 64 unique teams with Balanced Multiclass Accuracy of 0.505 (0.636 for the first team). The ODIN (Out-of-DIstribution detector for Neural networks) method used in the first approach was re-implemented using Keras. This is also my capstone project of Udacity Machine Learning Nanodegree. For more details please refer to the project report.
- Python 3.5 or above
- Keras 2.2.4
- TensorFlow 1.14
- pandas
- NumPy
- Matplotlib
- scikit-learn
- OpenCV-Python
- tqdm
- Augmentor: A modification of Augmentor 0.2.3 is under Augmentor
- Pillow-SIMD and libjpeg-turbo: Not necessary but recommend using them to improve image processing performance. I followed the fastai's document to install them.
| Diagnostic Category | Amount |
|---|---|
| Angiofibroma or fibrous papule | 1 |
| Angioma | 12 |
| Atypical melanocytic proliferation | 12 |
| Lentigo NOS | 70 |
| Lentigo simplex | 22 |
| Scar | 1 |
After downloading the dataset, all melanosis images can be retrieved from the dataset by using the notebook.
| Diagnostic Category | Amount |
|---|---|
| Melanosis | 16 |
- Put all data under a root folder like the tree structure shown below.
- ISIC_2019_Training_Input and ISIC_2019_Test_Input folders contain images of ISIC 2019 Training Data and Test Data respectively.
- Out_Distribution folder contains all out-of-distribution images from both ISIC Archive and Seven-Point datasets.
- ISIC_2019_Training_GroundTruth_DuplicateRemoved.csv is a copy of ISIC_2019_Training_GroundTruth.csv but removing two entries: ISIC_0067980 and ISIC_0069013.
RootFolder
│ ISIC_2019_Training_GroundTruth.csv
│ ISIC_2019_Training_GroundTruth_DuplicateRemoved.csv
│
└───ISIC_2019_Test_Input
│ │ ISIC_0034321.jpg
│ │ ISIC_0034322.jpg
│ │ ...
│ │
└───ISIC_2019_Training_Input
│ │ ISIC_0000000.jpg
│ │ ISIC_0000001.jpg
│ │ ...
│ │
└───Out_Distribution
│ │ 7pt_Fgl059.jpg
│ │ 7pt_Fhl002.jpg
│ │ ...
│ │ ISIC_0001115.jpg
│ │ ISIC_0001129.jpg
│ │ ...
Follow the steps starting from the Common Parameters cell in the notebook approach_1.ipynb and approach_2.ipynb to reproduce training and predicting processes of approach-1 and 2 respectively. You might need to change a few parameters according to the environment.
Here is an example code of training DenseNet201, Xception and ResNeXt50 models using approach-1. Assuming the path to the data root folder is /home.
python3 main.py /home --approach 1 --training --epoch 100 --batchsize 32 --maxqueuesize 10 --model DenseNet201 Xception ResNeXt50
Use -h to show usage messages.
python3 main.py -h
Please visit ISIC 2019 Challenge Leaderboards for complete rankings.
Team Rankings
Ranked 25th out of 64 unique teams with Balanced Multiclass Accuracy of 0.505 (0.636 for the first team)
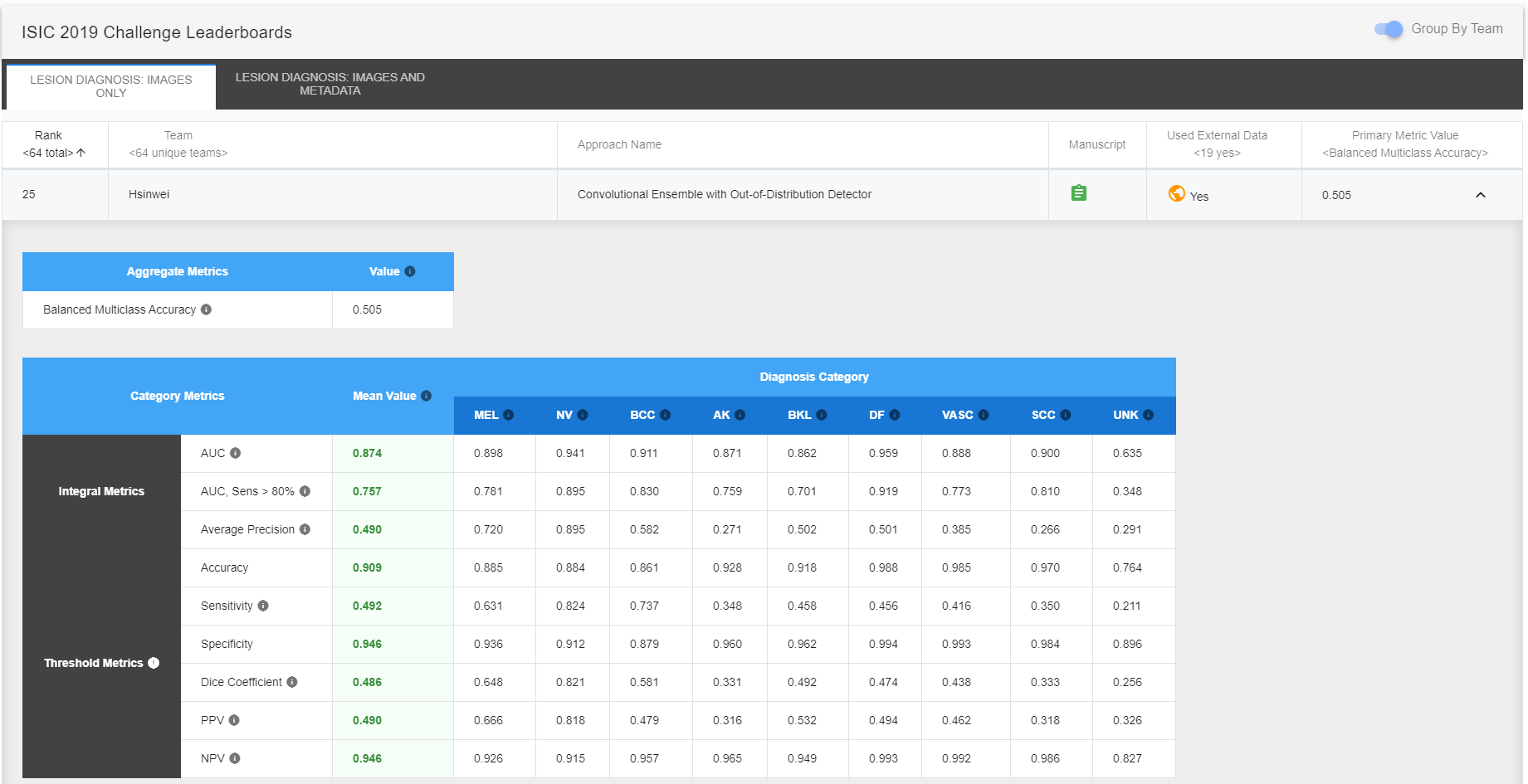
Approach Rankings
| Rank | Approach Name (129 approaches) | Balanced Multiclass Accuracy |
|---|---|---|
| 52 | Convolutional Ensemble with Out-of-Distribution Detector | 0.505 |
| 57 | Skin Lesion Classification using Ensemble of Convolutional Neural Networks | 0.499 |