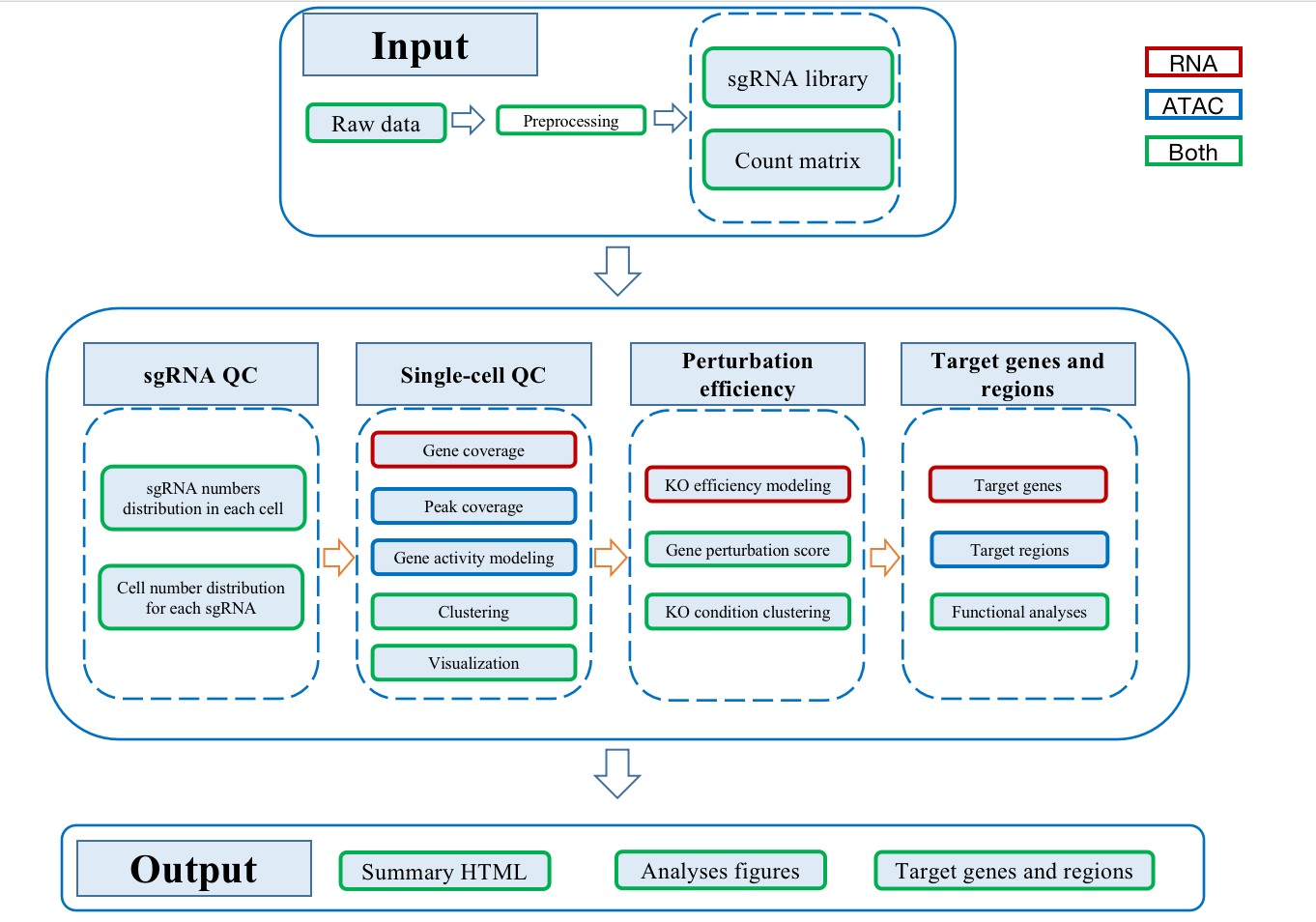
SCREE(Single-cell CRISPR scREen data analyses and pErturbation modeling) is a workflow for single-cell CRISPR screens data processing and analysis. SCREE has two separate section: pre-process and analysis. The pre-process section is to perform reads alignment and quantification based on cellranger and cellranger-atac. The analysis section is an R package including functions of sgRNA assignment, sgRNA information visualization, single-cell quality control, perturbation enrichment ratio calculation, perturbation efficiency evaluation, regulatory score estimation and downstream functional analysis. All the outputs of SCREE can be integrated into an html file.
We are hosting SCREEN documentation, instruction and tutorials at SCREE Website.
R >= 4.0.3
Seurat >= 4.0.3
cellranger >= 6.0
cellranger-atac
setuptools
devtools
Before installation and operation of SCREE, you should install cellranger (for scRNA-seq based data) and cellranger-atac (for scATAC-seq based data) first to make sure that SCREE can perform alignment and quantification accurately.
conda install -c hailinwei scree
If you only need the preprocess function, you can install SCREE via the pre-process/setup.py, which will not install the depend packages.
cd SCREE/pre-process
python setup.py install
$ R
> library(devtools)
> install_github("HailinWei98/SCREE")