The goal of stamp4r is to find diffrent features and plot extended errorbar plot.
You can install the development version of stamp4r like so:
devtools::install_github("wangzhichao1990/stamp4r)library(stamp4r)
## create a stamp object
obj <- stamp$new(feature_table = feature_table, metadata = metadata)
## cal diff
obj$cal_diff()
#> Joining with `by = join_by(SampleID)`
#> Warning in wilcox.test.default(x = DATA[[1L]], y = DATA[[2L]], ...):
#> 无法精確計算带连结的p值
#> Warning in wilcox.test.default(x = DATA[[1L]], y = DATA[[2L]], ...):
#> 无法精確計算带连结的置信区间
#> # A tibble: 23 × 9
#> Pathway estimate stati…¹ p.value conf.low conf.h…² method alter…³ p.adjust
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl>
#> 1 Energy M… 4.93e-3 1260 4.48e-16 3.85e-3 6.04e-3 Wilco… two.si… 1.03e-14
#> 2 Cell Com… 2.36e-5 1286 6.82e-13 -8.7 e-7 2.36e-5 Wilco… two.si… 7.85e-12
#> 3 Amino Ac… -6.88e-3 141 5.63e-10 -8.89e-3 -4.91e-3 Wilco… two.si… 4.32e- 9
#> 4 Metaboli… -1.92e-3 151 1.46e- 9 -2.47e-3 -1.37e-3 Wilco… two.si… 8.40e- 9
#> 5 Folding,… 2.02e-3 1078 3.49e- 7 1.25e-3 2.68e-3 Wilco… two.si… 1.61e- 6
#> 6 Glycan B… 1.91e-3 1051 2.20e- 6 1.13e-3 2.58e-3 Wilco… two.si… 8.43e- 6
#> 7 Transpor… -3.26e-4 267 8.67e- 6 -4.50e-4 -2.10e-4 Wilco… two.si… 2.85e- 5
#> 8 Metaboli… 1.35e-3 1022 1.31e- 5 7.59e-4 2.00e-3 Wilco… two.si… 3.77e- 5
#> 9 Membrane… -8.18e-3 315 1.22e- 4 -1.24e-2 -4.17e-3 Wilco… two.si… 3.11e- 4
#> 10 Metaboli… -9.37e-4 320 1.56e- 4 -1.50e-3 -4.94e-4 Wilco… two.si… 3.60e- 4
#> # … with 13 more rows, and abbreviated variable names ¹statistic, ²conf.high,
#> # ³alternative
obj$diff |> head()
#> # A tibble: 6 × 9
#> Pathway estimate stati…¹ p.value conf.low conf.h…² method alter…³ p.adjust
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <dbl>
#> 1 Energy Me… 4.93e-3 1260 4.48e-16 3.85e-3 6.04e-3 Wilco… two.si… 1.03e-14
#> 2 Cell Comm… 2.36e-5 1286 6.82e-13 -8.7 e-7 2.36e-5 Wilco… two.si… 7.85e-12
#> 3 Amino Aci… -6.88e-3 141 5.63e-10 -8.89e-3 -4.91e-3 Wilco… two.si… 4.32e- 9
#> 4 Metabolis… -1.92e-3 151 1.46e- 9 -2.47e-3 -1.37e-3 Wilco… two.si… 8.40e- 9
#> 5 Folding, … 2.02e-3 1078 3.49e- 7 1.25e-3 2.68e-3 Wilco… two.si… 1.61e- 6
#> 6 Glycan Bi… 1.91e-3 1051 2.20e- 6 1.13e-3 2.58e-3 Wilco… two.si… 8.43e- 6
#> # … with abbreviated variable names ¹statistic, ²conf.high, ³alternative
## filter diff features
obj$filter_diff_features()
#> [1] "Energy Metabolism"
#> [2] "Cell Communication"
#> [3] "Amino Acid Metabolism"
#> [4] "Metabolism of Other Amino Acids"
#> [5] "Folding, Sorting and Degradation"
#> [6] "Glycan Biosynthesis and Metabolism"
#> [7] "Transport and Catabolism"
#> [8] "Metabolism of Cofactors and Vitamins"
#> [9] "Membrane Transport"
#> [10] "Metabolism of Terpenoids and Polyketides"
#> [11] "Signal Transduction"
#> [12] "Cell Motility"
#> [13] "Signaling Molecules and Interaction"
#> [14] "Xenobiotics Biodegradation and Metabolism"
#> [15] "Enzyme Families"
#> [16] "Cell Growth and Death"
obj$diff_features
#> [1] "Energy Metabolism"
#> [2] "Cell Communication"
#> [3] "Amino Acid Metabolism"
#> [4] "Metabolism of Other Amino Acids"
#> [5] "Folding, Sorting and Degradation"
#> [6] "Glycan Biosynthesis and Metabolism"
#> [7] "Transport and Catabolism"
#> [8] "Metabolism of Cofactors and Vitamins"
#> [9] "Membrane Transport"
#> [10] "Metabolism of Terpenoids and Polyketides"
#> [11] "Signal Transduction"
#> [12] "Cell Motility"
#> [13] "Signaling Molecules and Interaction"
#> [14] "Xenobiotics Biodegradation and Metabolism"
#> [15] "Enzyme Families"
#> [16] "Cell Growth and Death"
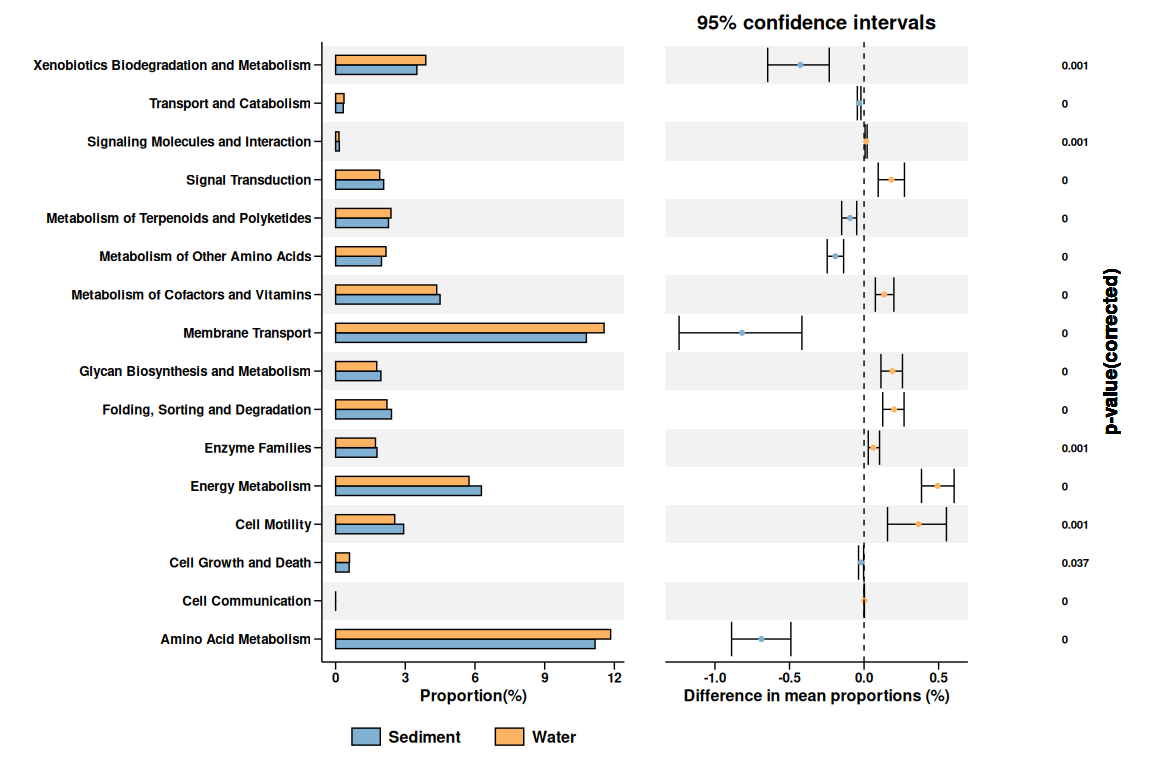
## extended errorbar plot
obj$plot_extended_errorbar()
#> Joining with `by = join_by(SampleID)`