It's based on "The Gaussian mixture probability hypothesis density filter" by Vo and Ma. Also, OSPA metric is provided for performance evaulation.
The gmphd.py contains two simulation examples.
The first is actually replica of the example in "Bayesian Multiple Target Filtering Using Random Finite Sets" by Vo, Vo, Clark used for performance evaulation of GMPHD filter. Targets lifes are based on Matlab code provided by Vo in http://ba-tuong.vo-au.com/codes.html and example doesn't contain target spawning.
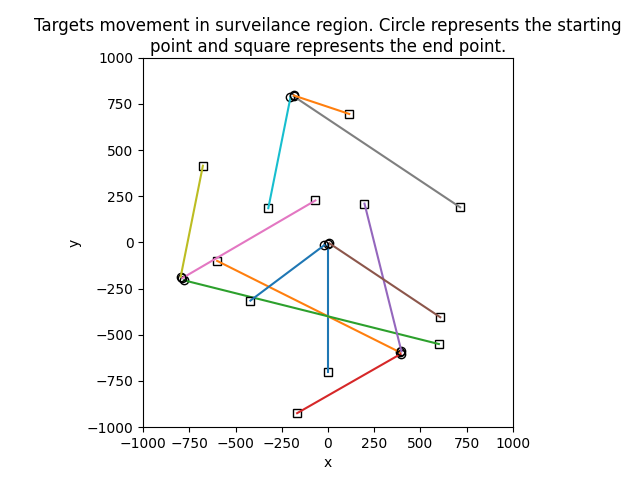 |
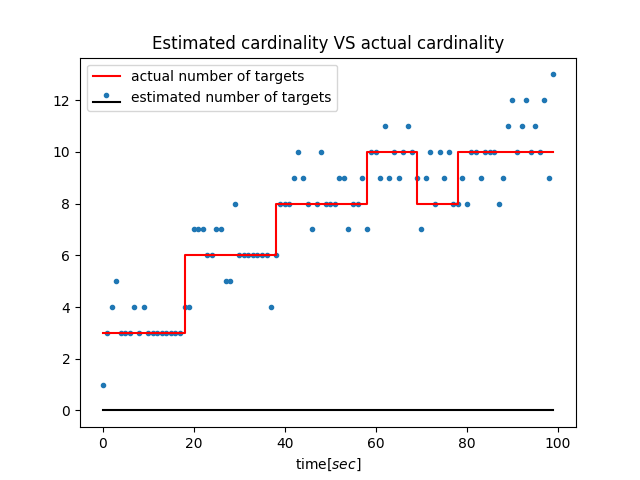 |
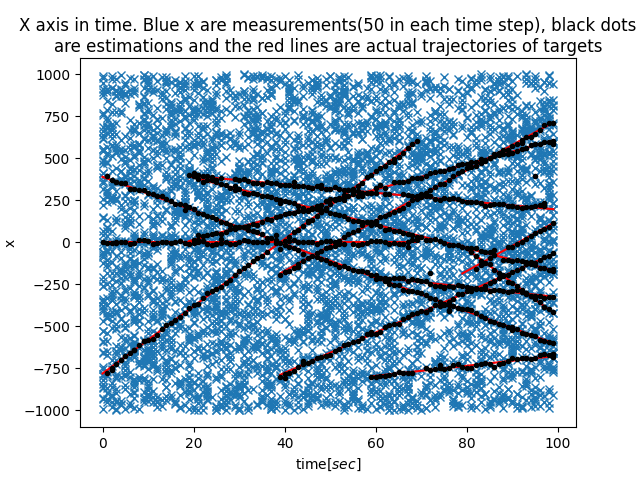 |
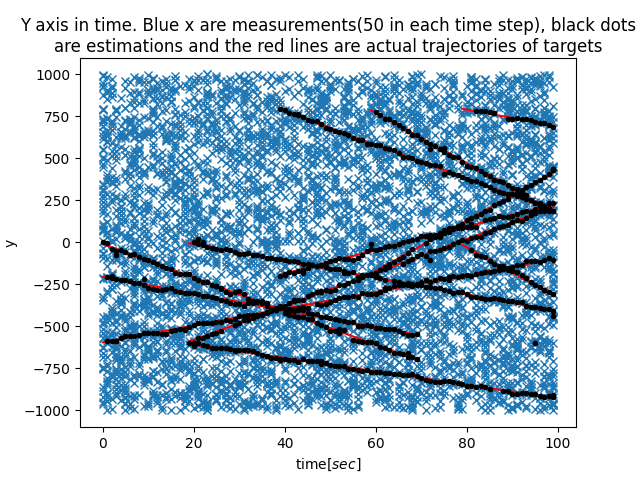 |
The second example is based on simulation example in "The Gaussian mixture probability hypothesis density filter" by Vo and Ma. Also, this example demonstrates the spawning situation.
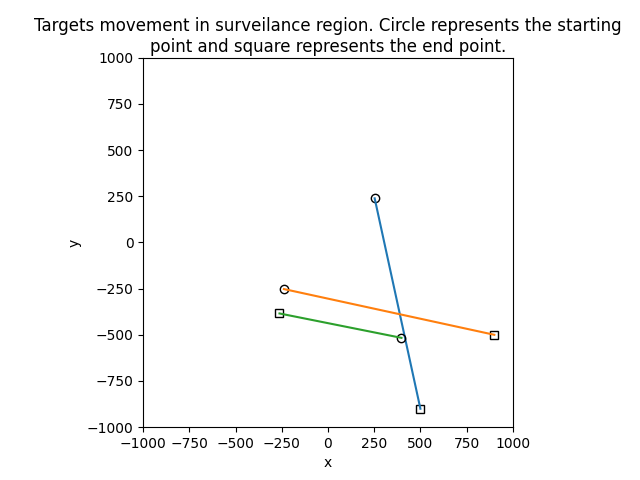 |
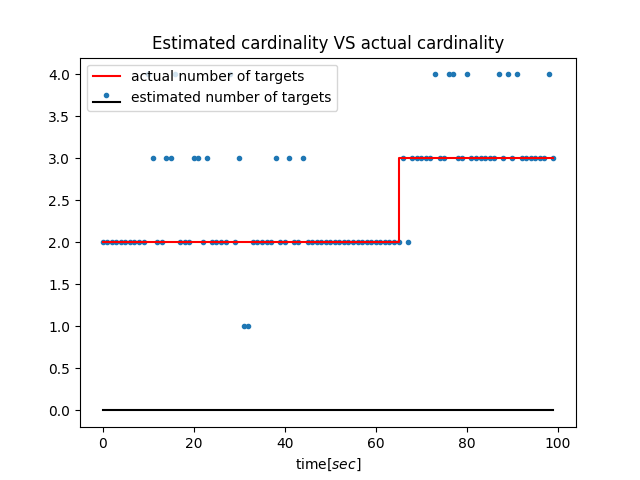 |
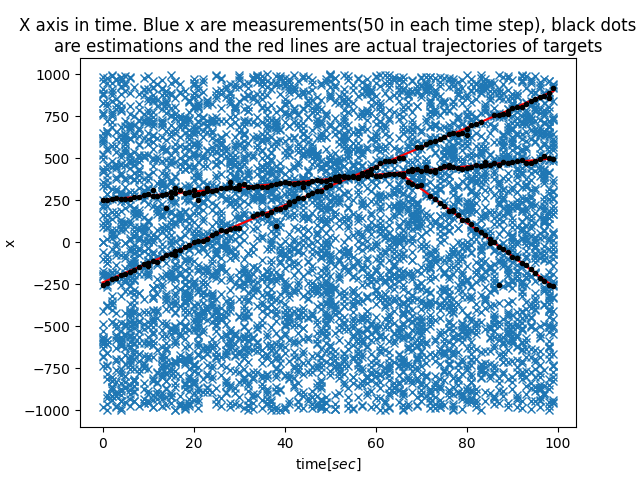 |
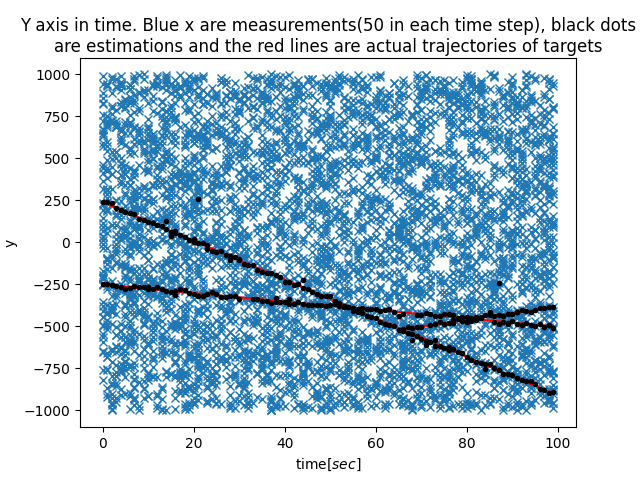 |
The simulation process steps are the following:
- Define the details of the process model:
model = process_model_for_example_1()- Create the description objects of the targets birth moment, death moment and starting points. Then generate trajectories based on the model and the given targets life description:
targets_birth_time, targets_death_time, targets_start = example1(model['num_scans'])
trajectories, targets_tracks = generate_trajectories(model, targets_birth_time, targets_death_time, targets_start,
noise=False)In the provided two simulation examples, targets are moving with constant velocity, we can make them move randomly in the
observation area by setting the noise parameter true.
- Generate measurements based on the target trajectories. Because in real situations sensor observations are noisy, in this step we just add the measurement noise for simulation purposes:
data = generate_measurements(model, trajectories)The data is list of collections of observations in each time step. For real filtering process, this fill be the
input from our sensors.
- Filter the measurement data with GMPHD filter. Filter also needs to know assumed process model, which is provided by creation of the filter object.
gmphd = GmphdFilter(model)
X_collection = gmphd.filter_data(data)All about the creation of the simulation process and running examples is in gmphd.py file. To switch between the two examples, just toggle comments at the beggining of the main function.
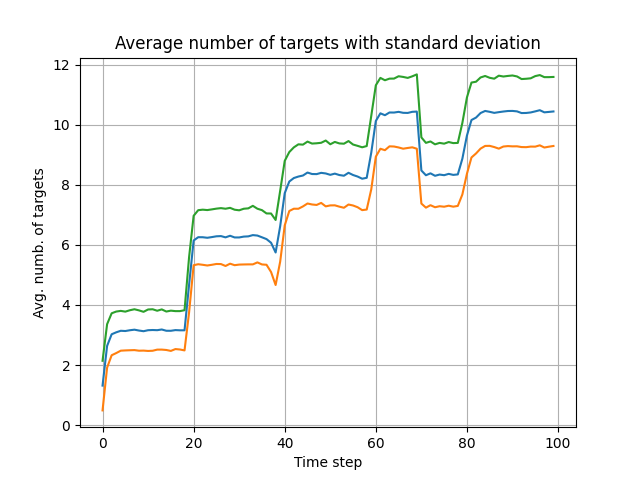
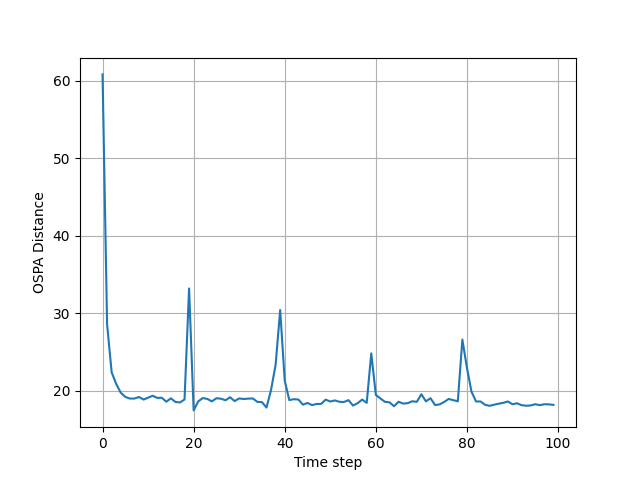
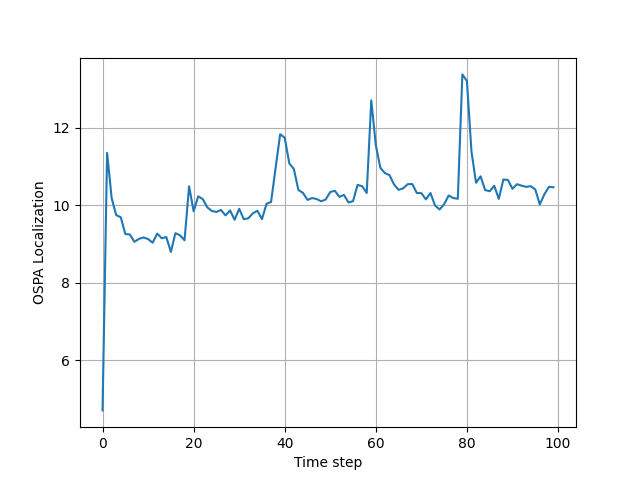
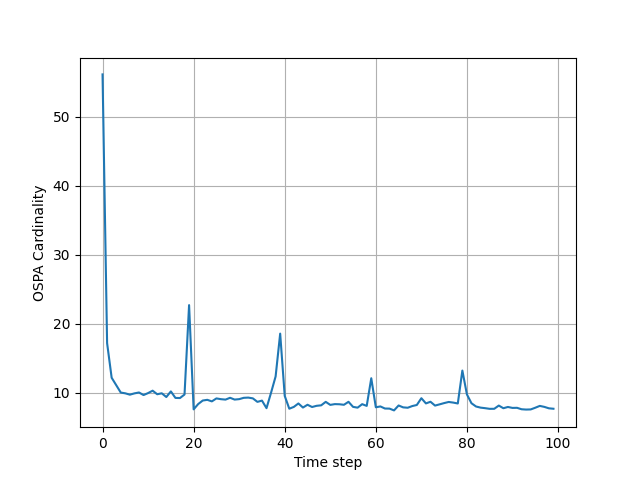
To measure the performance of the GMPHD filter, the averaged OSPA metric is used from "A consistent metric for performance evaluation of multi-object filters", by D. Schuhmacher, B.-T. Vo, and B.-N. Vo.
in monte_carlo_simulations_and_plots_of_results.py you can run
Monte Corle simulations, which is implemented in MC_run() function. These simulations can last quite long, and that is
why the result is saved in .pkl file. I have provided one such file with averaged 1000 MC simulations in
MC2ospatnum1000.pkl file, and you can plot it's result also in this file.
You can toggle between simulation and plotting the results just with commenting out the appropriate section.
 |
 |
 |
 |