Our web servise is available from https://orthoscope.jp.
If https://orthoscope.jp does not work, please try https://http://fish-evol.unit.oist.jp/orthoscope/ (8 Jan. 2019).
Also, mirror sites (http://fish-evol.org/orthoscope/) can be used (18 Mar. 2019).
Japanese instruction
Inoue J, Nakashima K, and Satoh N. ORTHOSCOPE analysis reveals the cellulose synthase gene in all tunicate genomes, but nowhere else in animal genomes. in prep.
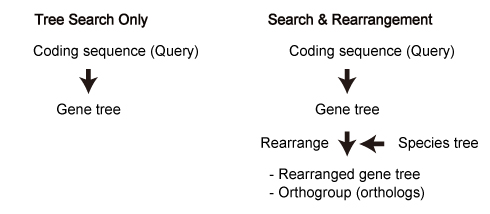
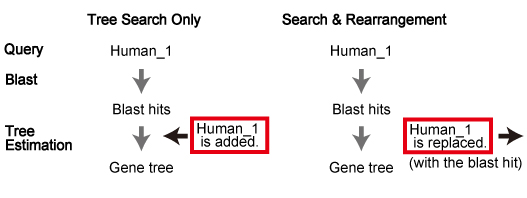
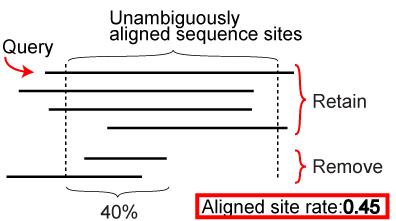
Queries. These sequences were used for "Tree Search Only" mode.
In this paper, maximum likelihood trees were estimated according to the process described in "Tree Estimation of Orthogroup Members (with Additional Sequences)". See below.
Inoue J. and Satoh N. 2019. ORTHOSCOPE: an automatic web tool of analytical pipeline for ortholog identification using a species tree. MBE in press.
| Actinopterygii | Vertebrata | Deuterostomia | Protostomia |
|---|---|---|---|
| PLCB1* | ALDH1A* | Brachyury | Brachyury |
| Queries | Queries | Queries | Queries |
| Result | Result | Result | Result |
From NCBI or Ensembl, query sequences can be downloaded.
For coding sequneces, please select CDS as follows.
- Download Coregonus lavaretus TSA file (GFIG00000000.1) form NCBI.
- Translate raw sequences into amino acid and coding sequences using TransDecoder.
./TransDecoder.LongOrfs -t GFIG01.1.fsa_nt
- Make blast databases using BLAST+.
makeblastdb -in longest_orfs.pep -dbtype prot -parse_seqids
makeblastdb -in longest_orfs.cds -dbtype nucl -parse_seqids
- BLASTP seaech against amino acid database.
blastp -query query.txt -db longest_orfs.pep -num_alignments 10 -evalue 1e-12 -out 010_out.txt
- Retrieve blast top hit sequences from coding sequence file using sequence id.
blastdbcmd -db longest_orfs.cds -dbtype nucl -entry_batch queryIDs.txt -out 020_out.txt
Coding sequence
Case 1: Query seqeunce is present in the ORTHOSCOPE database
Case 2: Query seqeunce is not present in the ORTHOSCOPE database
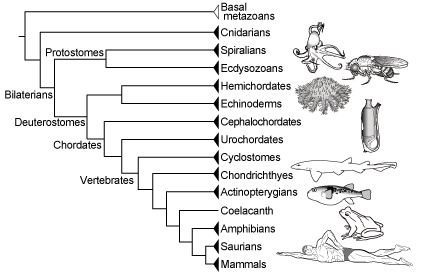
Our hypothetical species tree (newick) can be downloaded from here.
| Metazoa | Hexapoda | Urochordata | Vertebrata | Aves | Actinopterygii |
|---|
Phylogenetic relationships without references follow the NCBI Taxonomy Common Tree.
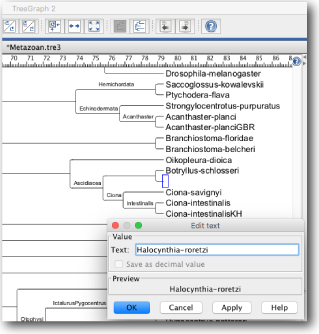
Newick formats can be modifed using TreeGraph2.
Dataset
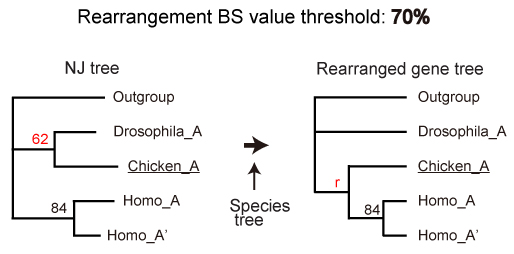
Rearrangement BS value threshold
NJ analysis is conducted using the software package Ape in R (coding) and FastME (amino acid). Rearrangement analysis is done using a method implemented in NOTUNG.
Feasibility of completion
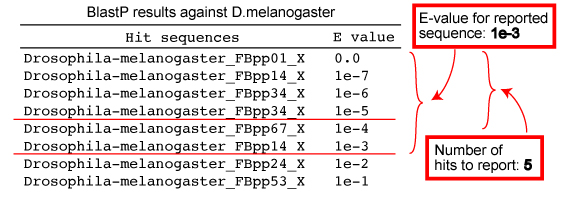
| Number of hits to report per genome | Number of species |
|---|---|
| 3 | <50 |
| 5 | <40 |
| 10 | <30 |
By using sequences of ORTHOSCOPE results, the analysis can be done on your own computer.
I made an analysis pipeline for this 2nd step. The script is specialized for a Macintosh use with Python 3. Windows users need some modifications.
Analysis pipeline with example data: DeuterostomeBra_2ndAnalysis.zip.
Estimation of the 2nd tree by the downloaded pipeline requires some dependencies to be installed and in the system path in your computer.
Available here: https://github.com/stamatak/standard-RAxML
Download the the latest release and extract it. Cd into the extracted directry (e.g., standard-RAxML-8.2.12), compile the PThreads version, and copy the executable to a directory in your system path, e.g.:
cd standard-RAxML-8.2.12
make -f Makefile.SSE3.PTHREADS.gcc
cp raxmlHPC-PTHREADS-SSE3 ~/bin
Add the address to your PATH. For example:
export PATH=$PATH:~/bin
Available here: https://mafft.cbrc.jp/alignment/software/.
After compilation, set your PATH following this site.
Available here: http://trimal.cgenomics.org/downloads.
Cd to trimAl/source, type make, and copy the executable.
make
cp trimal ~/bin
Available here: http://www.bork.embl.de/pal2nal/#Download.
Change the permission of perl script and copy it.
chmod 755 pal2nal.pl
cp pal2nal.pl ~/bin
R (3.5.2) is available from here.
By installing R, rscript will be installed automatically.
APE in R can be installed from the R console as follows:
install.packages("ape")
Using the downloaded pipeline, the 2nd gene trees will be estimated as follows:
- Based on the estimated rearranged NJ tree, users should select coding sequences of orthogroup and outgroups manually. Then the pipeline can start subsequent analyese.
- Selected sequences are aligned using MAFFT (Katoh et al. 2005).
- Multiple sequence alignments are trimmed by removing poorly aligned regions using TRIMAL 1.2 (Capella-Gutierrez et al. 2009) with the option “gappyout.”
- Corresponding cDNA sequences are forced onto the amino acid alignment using PAL2NAL (Suyama et al. 2006) to generate nucleotide alignments.
- Phylogenetic analysis is performed with RAxML 8.2.4 (Stamatakis et al. 2014), which invokes a rapid bootstrap analysis and searches for the best-scoring ML tree with the GTRGAMMA (Yang 1994a, 1994b) or GTRCAT model.
The actual rocess is as follows:
-
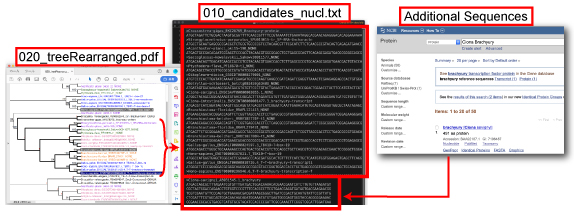
Decompress DeuterostomeBra_2ndAnalysis.zip. Open DeuterostomeBra_2ndAnalysis file and decompress 100_2ndTree.tar.gz file.
-
Select an appropriate outgroup and orthogroup members and save 010_candidates_nucl.txt file. The outgroup sequence should be placed at the top of alignment. Additional sequences can be included.
- Cd into 100_2ndTree directory.
- Run the pipeline.
./100_estimate2ndTree.py
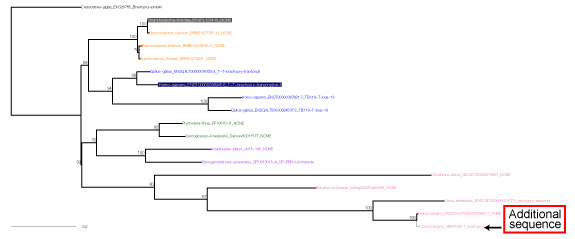
- ML tree is saved in 200_RAxMLtree_Exc3rd.pdf automatically.
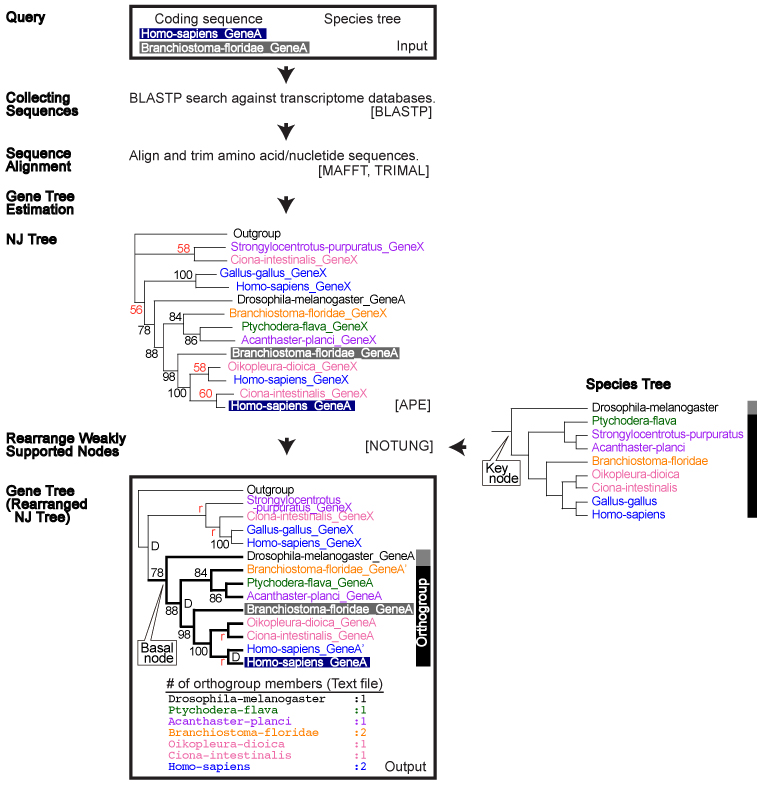
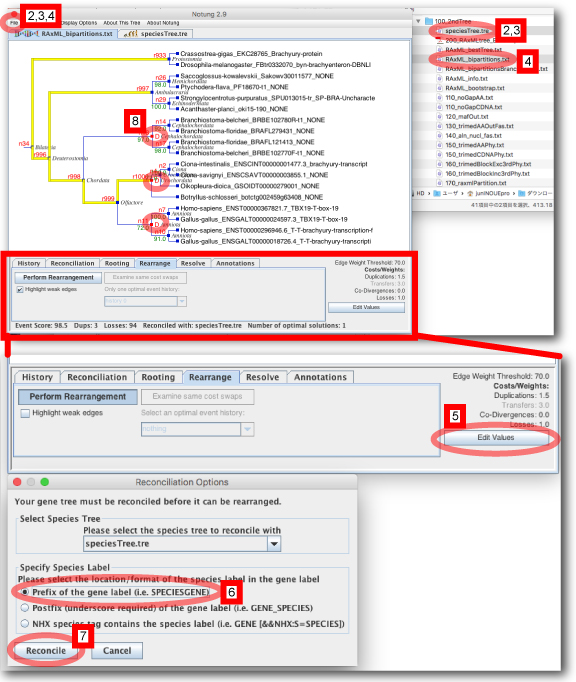
Using Notung, duplicated nodes can be identified. Here, we will analyze the gene tree of orthogroup members.
- Double click the downloaded .jar file (here, Notung-2.9.jar).
- Save the species tree (newick format) as a new file (here, speciesTree.tre), from 000_summary.txt file.
- Open the species tree file, speciesTree.tre (File > Open Gene Tree), from Notung.
- Open the gene tree file, RAxML_bootstrap.txt (File > Open Gene Tree).
- Set "Edge Weight THreshold" (here 70) from “Edit Values button“. This value corresponds to “Rearrangement BS value threshold” in ORTHOSCOPE.
- From "Rearrange" tab in the bottum, select "Prefix of the general label".
- Push "Reconcile" button.
- Duplicated nodes are shown with "D".
| Chrome | Firefox | Safari | IE |
|---|---|---|---|
| Supported | Supported | 11.0 or later | Not supported |
| Date | Version | Revision |
|---|---|---|
| 25 Jan. 2019 | Version 1.0.2 | Released. For Satoh et al. submitted, Data of Archaea, Plants, Bacteria, and Urochordata were newly added. |
| 21 Dec. 2018 | Version 1.0.1 | Released. In the rearranged gene tree, nodes identified as speciation events were marked with "D". |
| 18 Dec. 2018 | Version 1.0.1.beta | Xenacoelomorph, platyhelminth, priapulid, avian data were newly added. |
| 10 July 2018 | Version 1.0 | Published in Inoue and Satoh (2018). |
Available from here (10.5281/zenodo.2553737). 31 Jan. 2018.
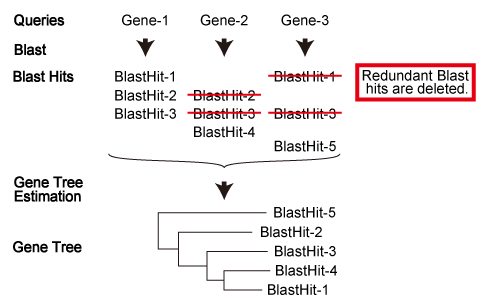
ORTHOSCOPE employs a genome-scale, protein-coding gene database (coding and amino acid sequence datasets) constructed for each species. In order to count numbers of orthologs in each species, only the longest sequence is used, when transcript variants exist for single locus.
Inoue J. and Satoh N. ORTHOSCOPE: An automatic web tool for phylogenetically inferring bilaterian orthogroups with user-selected taxa. Molecular Biology and Evolution, 36, 621–631. Link.
Previous versions:
Email: jun.inoue AT oist.jp